Degradation of p53 by human Alphapapillomavirus E6 proteins shows a stronger correlation with phylogeny than oncogenicity
- PMID: 20862247
- PMCID: PMC2941455
- DOI: 10.1371/journal.pone.0012816
Degradation of p53 by human Alphapapillomavirus E6 proteins shows a stronger correlation with phylogeny than oncogenicity
Abstract
Background: Human Papillomavirus (HPV) E6 induced p53 degradation is thought to be an essential activity by which high-risk human Alphapapillomaviruses (alpha-HPVs) contribute to cervical cancer development. However, most of our understanding is derived from the comparison of HPV16 and HPV11. These two viruses are relatively distinct viruses, making the extrapolation of these results difficult. In the present study, we expand the tested strains (types) to include members of all known HPV species groups within the Alphapapillomavirus genus.
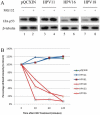
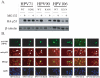
Principal findings: We report the biochemical activity of E6 proteins from 27 HPV types representing all alpha-HPV species groups to degrade p53 in human cells. Expression of E6 from all HPV types epidemiologically classified as group 1 carcinogens significantly reduced p53 levels. However, several types not associated with cancer (e.g., HPV53, HPV70 and HPV71) were equally active in degrading p53. HPV types within species groups alpha 5, 6, 7, 9 and 11 share a most recent common ancestor (MRCA) and all contain E6 ORFs that degrade p53. A unique exception, HPV71 E6 ORF that degraded p53 was outside this clade and is one of the most prevalent HPV types infecting the cervix in a population-based study of 10,000 women. Alignment of E6 ORFs identified an amino acid site that was highly correlated with the biochemical ability to degrade p53. Alteration of this amino acid in HPV71 E6 abrogated its ability to degrade p53, while alteration of this site in HPV71-related HPV90 and HPV106 E6s enhanced their capacity to degrade p53.
Conclusions: These data suggest that the alpha-HPV E6 proteins' ability to degrade p53 is an evolved phenotype inherited from a most recent common ancestor of the high-risk species that does not always segregate with carcinogenicity. In addition, we identified an amino-acid residue strongly correlated with viral p53 degrading potential.
Conflict of interest statement
Figures


Similar articles
-
p53 degradation activity, expression, and subcellular localization of E6 proteins from 29 human papillomavirus genotypes.J Virol. 2012 Jan;86(1):94-107. doi: 10.1128/JVI.00751-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013048 Free PMC article.
-
Activating transcription factor 3 activates p53 by preventing E6-associated protein from binding to E6.J Biol Chem. 2010 Apr 23;285(17):13201-10. doi: 10.1074/jbc.M109.058669. Epub 2010 Feb 18. J Biol Chem. 2010. PMID: 20167600 Free PMC article.
-
Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche.PLoS Pathog. 2015 Jun 18;11(6):e1004980. doi: 10.1371/journal.ppat.1004980. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26086730 Free PMC article.
-
Human papillomavirus genome variants.Virology. 2013 Oct;445(1-2):232-43. doi: 10.1016/j.virol.2013.07.018. Epub 2013 Aug 31. Virology. 2013. PMID: 23998342 Free PMC article. Review.
-
Hyperactivating p53 in Human Papillomavirus-Driven Cancers: A Potential Therapeutic Intervention.Mol Diagn Ther. 2022 May;26(3):301-308. doi: 10.1007/s40291-022-00583-5. Epub 2022 Apr 5. Mol Diagn Ther. 2022. PMID: 35380358 Free PMC article. Review.
Cited by
-
p53 degradation activity, expression, and subcellular localization of E6 proteins from 29 human papillomavirus genotypes.J Virol. 2012 Jan;86(1):94-107. doi: 10.1128/JVI.00751-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013048 Free PMC article.
-
Human Papillomavirus 11 Early Protein E6 Activates Autophagy by Repressing AKT/mTOR and Erk/mTOR.J Virol. 2019 May 29;93(12):e00172-19. doi: 10.1128/JVI.00172-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30971468 Free PMC article.
-
Papillomaviruses: Viral evolution, cancer and evolutionary medicine.Evol Med Public Health. 2015 Jan 28;2015(1):32-51. doi: 10.1093/emph/eov003. Evol Med Public Health. 2015. PMID: 25634317 Free PMC article. Review.
-
Papillomavirus E6 oncoproteins.Virology. 2013 Oct;445(1-2):115-37. doi: 10.1016/j.virol.2013.04.026. Epub 2013 May 24. Virology. 2013. PMID: 23711382 Free PMC article. Review.
-
Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.Nature. 2016 Jan 28;529(7587):541-5. doi: 10.1038/nature16481. Epub 2016 Jan 20. Nature. 2016. PMID: 26789255 Free PMC article.
References
-
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. - PubMed
-
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. - PubMed
-
- Parkin DM, Bray F. Chapter 2: The burden of HPV-related cancers. Vaccine. 2006;24(Suppl 3):S11–25. - PubMed
-
- Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

