Kinetic Modeling using BioPAX ontology
- PMID: 20862270
- PMCID: PMC2941992
- DOI: 10.1109/BIBM.2007.55
Kinetic Modeling using BioPAX ontology
Abstract
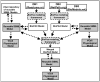
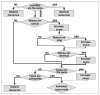
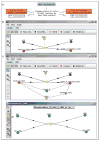
Thousands of biochemical interactions are available for download from curated databases such as Reactome, Pathway Interaction Database and other sources in the Biological Pathways Exchange (BioPAX) format. However, the BioPAX ontology does not encode the necessary information for kinetic modeling and simulation. The current standard for kinetic modeling is the System Biology Markup Language (SBML), but only a small number of models are available in SBML format in public repositories. Additionally, reusing and merging SBML models presents a significant challenge, because often each element has a value only in the context of the given model, and information encoding biological meaning is absent. We describe a software system that enables a variety of operations facilitating the use of BioPAX data to create kinetic models that can be visualized, edited, and simulated using the Virtual Cell (VCell), including improved conversion to SBML (for use with other simulation tools that support this format).
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
