Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer
- PMID: 20862303
- PMCID: PMC2940729
- DOI: 10.1371/journal.pcbi.1000933
Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer
Abstract
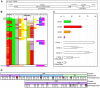
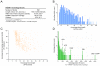
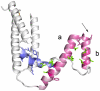
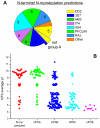
The var gene encoded hyper-variable Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) family mediates cytoadhesion of infected erythrocytes to human endothelium. Antibodies blocking cytoadhesion are important mediators of malaria immunity acquired by endemic populations. The development of a PfEMP1 based vaccine mimicking natural acquired immunity depends on a thorough understanding of the evolved PfEMP1 diversity, balancing antigenic variation against conserved receptor binding affinities. This study redefines and reclassifies the domains of PfEMP1 from seven genomes. Analysis of domains in 399 different PfEMP1 sequences allowed identification of several novel domain classes, and a high degree of PfEMP1 domain compositional order, including conserved domain cassettes not always associated with the established group A-E division of PfEMP1. A novel iterative homology block (HB) detection method was applied, allowing identification of 628 conserved minimal PfEMP1 building blocks, describing on average 83% of a PfEMP1 sequence. Using the HBs, similarities between domain classes were determined, and Duffy binding-like (DBL) domain subclasses were found in many cases to be hybrids of major domain classes. Related to this, a recombination hotspot was uncovered between DBL subdomains S2 and S3. The VarDom server is introduced, from which information on domain classes and homology blocks can be retrieved, and new sequences can be classified. Several conserved sequence elements were found, including: (1) residues conserved in all DBL domains predicted to interact and hold together the three DBL subdomains, (2) potential integrin binding sites in DBLα domains, (3) an acylation motif conserved in group A var genes suggesting N-terminal N-myristoylation, (4) PfEMP1 inter-domain regions proposed to be elastic disordered structures, and (5) several conserved predicted phosphorylation sites. Ideally, this comprehensive categorization of PfEMP1 will provide a platform for future studies on var/PfEMP1 expression and function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. - PubMed
-
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. - PubMed
-
- Staalsoe T, Shulman CE, Bulmer JN, Kawuondo K, Marsh K, et al. Variant surface antigen-specific IgG and protection against clinical consequences of pregnancy-associated Plasmodium falciparum malaria. Lancet. 2004;363:283–289. - PubMed
-
- Marsh K, Howard RJ. Antigens induced on erythrocytes by P. falciparum: expression of diverse and conserved determinants. Science. 1986;231:150–153. - PubMed
-
- Fried M, Nosten F, Brockman A, Brabin BJ, Duffy PE. Maternal antibodies block malaria. Nature. 1998;395:851–852. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

