Comparative Genomic Hybridization (CGH) reveals a neo-X chromosome and biased gene movement in stalk-eyed flies (genus Teleopsis)
- PMID: 20862308
- PMCID: PMC2940734
- DOI: 10.1371/journal.pgen.1001121
Comparative Genomic Hybridization (CGH) reveals a neo-X chromosome and biased gene movement in stalk-eyed flies (genus Teleopsis)
Abstract
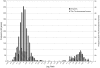
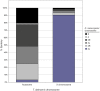
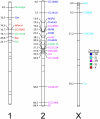
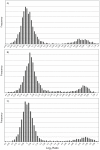
Chromosomal location has a significant effect on the evolutionary dynamics of genes involved in sexual dimorphism, impacting both the pattern of sex-specific gene expression and the rate of duplication and protein evolution for these genes. For nearly all non-model organisms, however, knowledge of chromosomal gene content is minimal and difficult to obtain on a genomic scale. In this study, we utilized Comparative Genomic Hybridization (CGH), using probes designed from EST sequence, to identify genes located on the X chromosome of four species in the stalk-eyed fly genus Teleopsis. Analysis of log(2) ratio values of female-to-male hybridization intensities from the CGH microarrays for over 3,400 genes reveals a strongly bimodal distribution that clearly differentiates autosomal from X-linked genes for all four species. Genotyping of 33 and linkage mapping of 28 of these genes in Teleopsis dalmanni indicate the CGH results correctly identified chromosomal location in all cases. Syntenic comparison with Drosophila indicates that 90% of the X-linked genes in Teleopsis are homologous to genes located on chromosome 2L in Drosophila melanogaster, suggesting the formation of a nearly complete neo-X chromosome from Muller element B in the dipteran lineage leading to Teleopsis. Analysis of gene movement both relative to Drosophila and within Teleopsis indicates that gene movement is significantly associated with 1) rates of protein evolution, 2) the pattern of gene duplication, and 3) the evolution of eyespan sexual dimorphism. Overall, this study reveals that diopsids are a critical group for understanding the evolution of sex chromosomes within Diptera. In addition, we demonstrate that CGH is a useful technique for identifying chromosomal sex-linkage and should be applicable to other organisms with EST or partial genomic information.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Genomic analysis of a sexually-selected character: EST sequencing and microarray analysis of eye-antennal imaginal discs in the stalk-eyed fly Teleopsis dalmanni (Diopsidae).BMC Genomics. 2009 Aug 5;10:361. doi: 10.1186/1471-2164-10-361. BMC Genomics. 2009. PMID: 19656405 Free PMC article.
-
Germline transformation of the stalk-eyed fly, Teleopsis dalmanni.BMC Mol Biol. 2010 Nov 16;11:86. doi: 10.1186/1471-2199-11-86. BMC Mol Biol. 2010. PMID: 21080934 Free PMC article.
-
Length polymorphism and head shape association among genes with polyglutamine repeats in the stalk-eyed fly, Teleopsis dalmanni.BMC Evol Biol. 2010 Jul 27;10:227. doi: 10.1186/1471-2148-10-227. BMC Evol Biol. 2010. PMID: 20663190 Free PMC article.
-
Gene duplication, tissue-specific gene expression and sexual conflict in stalk-eyed flies (Diopsidae).Philos Trans R Soc Lond B Biol Sci. 2012 Aug 19;367(1600):2357-75. doi: 10.1098/rstb.2011.0287. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22777023 Free PMC article. Review.
-
Stalk-eyed flies (Diopsidae): modelling the evolution and development of an exaggerated sexual trait.Bioessays. 2007 Mar;29(3):300-7. doi: 10.1002/bies.20543. Bioessays. 2007. PMID: 17295307 Review.
Cited by
-
Sex Chromosome Evolution in Muscid Flies.G3 (Bethesda). 2020 Apr 9;10(4):1341-1352. doi: 10.1534/g3.119.400923. G3 (Bethesda). 2020. PMID: 32051221 Free PMC article.
-
The X chromosome of the German cockroach, Blattella germanica, is homologous to a fly X chromosome despite 400 million years divergence.BMC Biol. 2019 Dec 5;17(1):100. doi: 10.1186/s12915-019-0721-x. BMC Biol. 2019. PMID: 31806031 Free PMC article.
-
Evolution of multiple sex-chromosomes associated with dynamic genome reshuffling in Leptidea wood-white butterflies.Heredity (Edinb). 2020 Sep;125(3):138-154. doi: 10.1038/s41437-020-0325-9. Epub 2020 Jun 9. Heredity (Edinb). 2020. PMID: 32518391 Free PMC article.
-
The house fly Y Chromosome is young and minimally differentiated from its ancient X Chromosome partner.Genome Res. 2017 Aug;27(8):1417-1426. doi: 10.1101/gr.215509.116. Epub 2017 Jun 15. Genome Res. 2017. PMID: 28619849 Free PMC article.
-
Unraveling the complex evolutionary history of lepidopteran chromosomes through ancestral chromosome reconstruction and novel chromosome nomenclature.BMC Biol. 2023 Nov 20;21(1):265. doi: 10.1186/s12915-023-01762-4. BMC Biol. 2023. PMID: 37981687 Free PMC article.
References
-
- Fisher RA. The evolution of dominance. Biol Rev. 1931;6:345–368.
-
- Muller HJ. Some genetic aspects of sex. Am Nat. 1932;66:118–138.
-
- Bull JJ. Menlo Park: Benjamin Cummings; 1983. Evolution of Sex Determining Mechanisms.
-
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. - PubMed
-
- White MJD. Cambridge: Cambridge University Press; 1973. Animal Cytology and Evolution.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

