Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom
- PMID: 20863387
- PMCID: PMC2997006
- DOI: 10.1186/1471-2164-11-510
Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom
Abstract
Background: The nutrient-sensing Tor pathway governs cell growth and is conserved in nearly all eukaryotic organisms from unicellular yeasts to multicellular organisms, including humans. Tor is the target of the immunosuppressive drug rapamycin, which in complex with the prolyl isomerase FKBP12 inhibits Tor functions. Rapamycin is a gold standard drug for organ transplant recipients that was approved by the FDA in 1999 and is finding additional clinical indications as a chemotherapeutic and antiproliferative agent. Capitalizing on the plethora of recently sequenced genomes we have conducted comparative genomic studies to annotate the Tor pathway throughout the fungal kingdom and related unicellular opisthokonts, including Monosiga brevicollis, Salpingoeca rosetta, and Capsaspora owczarzaki.
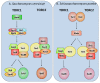
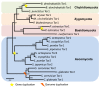
Results: Interestingly, the Tor signaling cascade is absent in three microsporidian species with available genome sequences, the only known instance of a eukaryotic group lacking this conserved pathway. The microsporidia are obligate intracellular pathogens with highly reduced genomes, and we hypothesize that they lost the Tor pathway as they adapted and streamlined their genomes for intracellular growth in a nutrient-rich environment. Two TOR paralogs are present in several fungal species as a result of either a whole genome duplication or independent gene/segmental duplication events. One such event was identified in the amphibian pathogen Batrachochytrium dendrobatidis, a chytrid responsible for worldwide global amphibian declines and extinctions.
Conclusions: The repeated independent duplications of the TOR gene in the fungal kingdom might reflect selective pressure acting upon this kinase that populates two proteinaceous complexes with different cellular roles. These comparative genomic analyses illustrate the evolutionary trajectory of a central nutrient-sensing cascade that enables diverse eukaryotic organisms to respond to their natural environments.
Figures



Similar articles
-
Evolution of the sex-related locus and genomic features shared in microsporidia and fungi.PLoS One. 2010 May 7;5(5):e10539. doi: 10.1371/journal.pone.0010539. PLoS One. 2010. PMID: 20479876 Free PMC article.
-
Analysis of septins across kingdoms reveals orthology and new motifs.BMC Evol Biol. 2007 Jul 1;7:103. doi: 10.1186/1471-2148-7-103. BMC Evol Biol. 2007. PMID: 17601340 Free PMC article.
-
In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins.Mol Biol Evol. 2012 Nov;29(11):3371-84. doi: 10.1093/molbev/mss143. Epub 2012 May 23. Mol Biol Evol. 2012. PMID: 22628532
-
The intriguing nature of microsporidian genomes.Brief Funct Genomics. 2011 May;10(3):115-24. doi: 10.1093/bfgp/elq032. Epub 2010 Dec 21. Brief Funct Genomics. 2011. PMID: 21177329 Review.
-
Evolutionary relationships between Saccharomyces cerevisiae and other fungal species as determined from genome comparisons.Rev Iberoam Micol. 2005 Dec;22(4):217-22. doi: 10.1016/s1130-1406(05)70046-2. Rev Iberoam Micol. 2005. PMID: 16499414 Review.
Cited by
-
Selection of Orthologous Genes for Construction of a Highly Resolved Phylogenetic Tree and Clarification of the Phylogeny of Trichosporonales Species.PLoS One. 2015 Aug 4;10(8):e0131217. doi: 10.1371/journal.pone.0131217. eCollection 2015. PLoS One. 2015. PMID: 26241762 Free PMC article.
-
Vitellogenins - Yolk Gene Function and Regulation in Caenorhabditis elegans.Front Physiol. 2019 Aug 21;10:1067. doi: 10.3389/fphys.2019.01067. eCollection 2019. Front Physiol. 2019. PMID: 31551797 Free PMC article. Review.
-
Cross-Talk and Multiple Control of Target of Rapamycin (TOR) in Sclerotinia sclerotiorum.Microbiol Spectr. 2023 Mar 21;11(2):e0001323. doi: 10.1128/spectrum.00013-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 36943069 Free PMC article.
-
Hormetic Effect of H2O2 in Saccharomyces cerevisiae: Involvement of TOR and Glutathione Reductase.Dose Response. 2016 Mar 30;14(2):1559325816636130. doi: 10.1177/1559325816636130. eCollection 2016 Apr-Jun. Dose Response. 2016. PMID: 27099601 Free PMC article.
-
Constitutive activation of TORC1 signalling attenuates virulence in the cross-kingdom fungal pathogen Fusarium oxysporum.Mol Plant Pathol. 2023 Apr;24(4):289-301. doi: 10.1111/mpp.13292. Epub 2023 Feb 24. Mol Plant Pathol. 2023. PMID: 36840362 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

