Self-association of the Lentivirus protein, Nef
- PMID: 20863404
- PMCID: PMC2955668
- DOI: 10.1186/1742-4690-7-77
Self-association of the Lentivirus protein, Nef
Abstract
Background: The HIV-1 pathogenic factor, Nef, is a multifunctional protein present in the cytosol and on membranes of infected cells. It has been proposed that a spatial and temporal regulation of the conformation of Nef sequentially matches Nef's multiple functions to the process of virion production. Further, it has been suggested that dimerization is required for multiple Nef activities. A dimerization interface has been proposed based on intermolecular contacts between Nefs within hexagonal Nef/FynSH3 crystals. The proposed dimerization interface consists of the hydrophobic B-helix and flanking salt bridges between R105 and D123. Here, we test whether Nef self-association is mediated by this interface and address the overall significance of oligomerization.
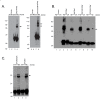
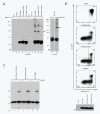
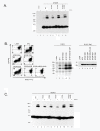
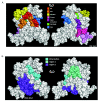
Results: By co-immunoprecipitation assays, we demonstrated that HIV-1Nef exists as monomers and oligomers with about half of the Nef protomers oligomerized. Nef oligomers were found to be present in the cytosol and on membranes. Removal of the myristate did not enhance the oligomerization of soluble Nef. Also, SIVNef oligomerizes despite lacking a dimerization interface functionally homologous to that proposed for HIV-1Nef. Moreover, HIV-1Nef and SIVNef form hetero-oligomers demonstrating the existence of homologous oligomerization interfaces that are distinct from that previously proposed (R105-D123). Intracellular cross-linking by formaldehyde confirmed that SF2Nef dimers are present in intact cells, but surprisingly self-association was dependent on R105, but not D123. SIV(MAC239)Nef can be cross-linked at its only cysteine, C55, and SF2Nef is also cross-linked, but at C206 instead of C55, suggesting that Nefs exhibit multiple dimeric structures. ClusPro dimerization analysis of HIV-1Nef homodimers and HIV-1Nef/SIVNef heterodimers identified a new potential dimerization interface, including a dibasic motif at R105-R106 and a six amino acid hydrophobic surface.
Conclusions: We have demonstrated significant levels of intracellular Nef oligomers by immunoprecipitation from cellular extracts. However, our results are contrary to the identification of salt bridges between R105 and D123 as necessary for self-association. Importantly, binding between HIV-1Nef and SIVNef demonstrates evolutionary conservation and therefore significant function(s) for oligomerization. Based on modeling studies of Nef self-association, we propose a new dimerization interface. Finally, our findings support a stochastic model of Nef function with a dispersed intracellular distribution of Nef oligomers.
Figures





Similar articles
-
HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication.J Mol Biol. 2009 Nov 27;394(2):329-42. doi: 10.1016/j.jmb.2009.09.047. Epub 2009 Sep 23. J Mol Biol. 2009. PMID: 19781555 Free PMC article.
-
Nef homodimers down-regulate SERINC5 by AP-2-mediated endocytosis to promote HIV-1 infectivity.J Biol Chem. 2020 Nov 13;295(46):15540-15552. doi: 10.1074/jbc.RA120.014668. Epub 2020 Sep 1. J Biol Chem. 2020. PMID: 32873704 Free PMC article.
-
A single β-octyl glucoside molecule induces HIV-1 Nef dimer formation in the absence of partner protein binding.PLoS One. 2018 Feb 7;13(2):e0192512. doi: 10.1371/journal.pone.0192512. eCollection 2018. PLoS One. 2018. PMID: 29415006 Free PMC article.
-
How HIV Nef Proteins Hijack Membrane Traffic To Promote Infection.J Virol. 2019 Nov 26;93(24):e01322-19. doi: 10.1128/JVI.01322-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31578291 Free PMC article. Review.
-
Structure, function, and inhibitor targeting of HIV-1 Nef-effector kinase complexes.J Biol Chem. 2020 Oct 30;295(44):15158-15171. doi: 10.1074/jbc.REV120.012317. Epub 2020 Aug 29. J Biol Chem. 2020. PMID: 32862141 Free PMC article. Review.
Cited by
-
Consequences of HLA-B*13-Associated Escape Mutations on HIV-1 Replication and Nef Function.J Virol. 2015 Nov;89(22):11557-71. doi: 10.1128/JVI.01955-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355081 Free PMC article.
-
Brain transcriptome-wide screen for HIV-1 Nef protein interaction partners reveals various membrane-associated proteins.PLoS One. 2012;7(12):e51578. doi: 10.1371/journal.pone.0051578. Epub 2012 Dec 17. PLoS One. 2012. PMID: 23284715 Free PMC article.
-
HIV-1 Nef Targets HDAC6 to Assure Viral Production and Virus Infection.Front Microbiol. 2019 Oct 30;10:2437. doi: 10.3389/fmicb.2019.02437. eCollection 2019. Front Microbiol. 2019. PMID: 31736889 Free PMC article.
-
The Nef-infectivity enigma: mechanisms of enhanced lentiviral infection.Curr HIV Res. 2011 Oct;9(7):474-89. doi: 10.2174/157016211798842099. Curr HIV Res. 2011. PMID: 22103831 Free PMC article. Review.
-
Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function.Virology. 2011 Nov 10;420(1):51-65. doi: 10.1016/j.virol.2011.08.015. Epub 2011 Sep 13. Virology. 2011. PMID: 21920573 Free PMC article.
References
-
- Foster JL, Garcia JV. Role of Nef in HIV-1 replication and pathogenesis. Adv Pharmacol. 2007;55:389–409. full_text. - PubMed
-
- Gorry PR, McPhee DA, Verity E, Dyer WB, Wesselingh SL, Learmont J, Sullivan JS, Roche M, Zaunders JJ, Gabuzda D, Crowe SM, Mills J, Lewin SR, Brew BJ, Cunningham AL, Churchill MJ. Pathogenicity and immunogenicity of attenuated, nef-deleted HIV-1 strains in vivo. Retrovirology. 2007;4:66. doi: 10.1186/1742-4690-4-66. - DOI - PMC - PubMed
-
- Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5(10):1361–1372. doi: 10.1016/S0969-2126(97)00286-4. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

