HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II
- PMID: 20864038
- PMCID: PMC4333224
- DOI: 10.1016/j.molcel.2010.08.023
HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II
Abstract
RNA polymerases are key multisubunit cellular enzymes. Microscopy studies indicated that RNA polymerase I assembles near its promoter. However, the mechanism by which RNA polymerase II is assembled from its 12 subunits remains unclear. We show here that RNA polymerase II subunits Rpb1 and Rpb3 accumulate in the cytoplasm when assembly is prevented and that nuclear import of Rpb1 requires the presence of all subunits. Using MS-based quantitative proteomics, we characterized assembly intermediates. These included a cytoplasmic complex containing subunits Rpb1 and Rpb8 associated with the HSP90 cochaperone hSpagh (RPAP3) and the R2TP/Prefoldin-like complex. Remarkably, HSP90 activity stabilized incompletely assembled Rpb1 in the cytoplasm. Our data indicate that RNA polymerase II is built in the cytoplasm and reveal quality-control mechanisms that link HSP90 to the nuclear import of fully assembled enzymes. hSpagh also bound the free RPA194 subunit of RNA polymerase I, suggesting a general role in assembling RNA polymerases.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

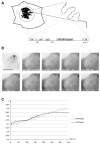

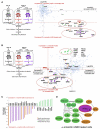


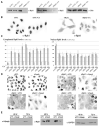
References
-
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
