The 5'-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway
- PMID: 20864040
- PMCID: PMC2945613
- DOI: 10.1016/j.molcel.2010.08.021
The 5'-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway
Abstract
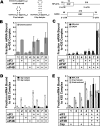
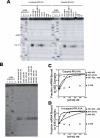
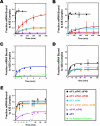
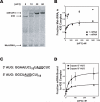
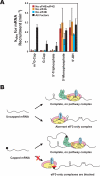
Translational control is frequently exerted at the stage of mRNA recruitment to the initiating ribosome. We have reconstituted mRNA recruitment to the 43S preinitiation complex (PIC) using purified S. cerevisiae components. We show that eIF3 and the eIF4 factors not only stabilize binding of mRNA to the PIC, they also dramatically increase the rate of recruitment. Although capped mRNAs require eIF3 and the eIF4 factors for efficient recruitment to the PIC, uncapped mRNAs can be recruited in the presence of eIF3 alone. The cap strongly inhibits this alternative recruitment pathway, imposing a requirement for the eIF4 factors for rapid and stable binding of natural mRNA. Our data suggest that the 5' cap serves as both a positive and negative element in mRNA recruitment, promoting initiation in the presence of the canonical group of mRNA handling factors while preventing binding to the ribosome via an aberrant, alternative pathway requiring only eIF3.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

