Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins
- PMID: 20865010
- PMCID: PMC4068012
- DOI: 10.1038/nrc2931
Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins
Abstract
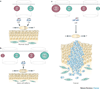
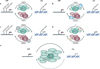
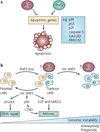
The discovery that cancer can be governed above and beyond the level of our DNA presents a new era for designing therapies that reverse the epigenetic state of a tumour cell. Understanding how altered chromatin dynamics leads to malignancy is essential for controlling tumour cells while sparing normal cells. Polycomb and trithorax group proteins are evolutionarily conserved and maintain chromatin in the 'off' or 'on' states, thereby preventing or promoting gene expression, respectively. Recent work highlights the dynamic interplay between these opposing classes of proteins, providing new avenues for understanding how these epigenetic regulators function in tumorigenesis.
Figures



References
-
- Wang JCa, D JE. Cancer stem cells: Lessons from leukemia. Trends Cell Biol. 2005;15:494–501. - PubMed
-
- Clarke MF, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. - PubMed
-
- Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. - PubMed
-
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nature Reviews Cancer. 2003;3:895–902. - PubMed
-
- Hill RP. Identifying cancer stem cells in solid tumors: case not proven. Cancer Res. 2006;66:1891–1895. discussion 1890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

