Genetic analysis of baker's yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability
- PMID: 20865162
- PMCID: PMC2928781
- DOI: 10.1371/journal.pgen.1001083
Genetic analysis of baker's yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability
Abstract
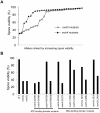
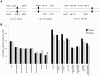
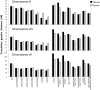
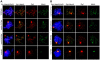
During meiosis, the Msh4-Msh5 complex is thought to stabilize single-end invasion intermediates that form during early stages of recombination and subsequently bind to Holliday junctions to facilitate crossover formation. To analyze Msh4-Msh5 function, we mutagenized 57 residues in Saccharomyces cerevisiae Msh4 and Msh5 that are either conserved across all Msh4/5 family members or are specific to Msh4 and Msh5. The Msh5 subunit appeared more sensitive to mutagenesis. We identified msh4 and msh5 threshold (msh4/5-t) mutants that showed wild-type spore viability and crossover interference but displayed, compared to wild-type, up to a two-fold decrease in crossing over on large and medium sized chromosomes (XV, VII, VIII). Crossing over on a small chromosome, however, approached wild-type levels. The msh4/5-t mutants also displayed synaptonemal complex assembly defects. A triple mutant containing a msh4/5-t allele and mutations that decreased meiotic double-strand break levels (spo11-HA) and crossover interference (pch2Δ) showed synergistic defects in spore viability. Together these results indicate that the baker's yeast meiotic cell does not require the ∼90 crossovers maintained by crossover homeostasis to form viable spores. They also show that Pch2-mediated crossover interference is important to maintain meiotic viability when crossovers become limiting.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The pch2Delta mutation in baker's yeast alters meiotic crossover levels and confers a defect in crossover interference.PLoS Genet. 2009 Jul;5(7):e1000571. doi: 10.1371/journal.pgen.1000571. Epub 2009 Jul 24. PLoS Genet. 2009. PMID: 19629178 Free PMC article.
-
Regulation of Msh4-Msh5 association with meiotic chromosomes in budding yeast.Genetics. 2021 Oct 2;219(2):iyab102. doi: 10.1093/genetics/iyab102. Genetics. 2021. PMID: 34849874 Free PMC article.
-
Variation in crossover frequencies perturb crossover assurance without affecting meiotic chromosome segregation in Saccharomyces cerevisiae.Genetics. 2015 Feb;199(2):399-412. doi: 10.1534/genetics.114.172320. Epub 2014 Dec 2. Genetics. 2015. PMID: 25467183 Free PMC article.
-
Roles for mismatch repair family proteins in promoting meiotic crossing over.DNA Repair (Amst). 2016 Feb;38:84-93. doi: 10.1016/j.dnarep.2015.11.024. Epub 2015 Dec 2. DNA Repair (Amst). 2016. PMID: 26686657 Free PMC article. Review.
-
[Homologs of MutS and MutL during mammalian meiosis].Med Sci (Paris). 2003 Jan;19(1):85-91. doi: 10.1051/medsci/200319185. Med Sci (Paris). 2003. PMID: 12836196 Review. French.
Cited by
-
Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction.PLoS Genet. 2012 Jul;8(7):e1002810. doi: 10.1371/journal.pgen.1002810. Epub 2012 Jul 5. PLoS Genet. 2012. PMID: 22792079 Free PMC article.
-
Population Genomics Reveals Speciation and Introgression between Brown Norway Rats and Their Sibling Species.Mol Biol Evol. 2017 Sep 1;34(9):2214-2228. doi: 10.1093/molbev/msx157. Mol Biol Evol. 2017. PMID: 28482038 Free PMC article.
-
A Zip3-like protein plays a role in crossover formation in the SC-less meiosis of the protist Tetrahymena.Mol Biol Cell. 2017 Mar 15;28(6):825-833. doi: 10.1091/mbc.E16-09-0678. Epub 2017 Jan 18. Mol Biol Cell. 2017. PMID: 28100637 Free PMC article.
-
Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae.Genetics. 2011 Oct;189(2):423-39. doi: 10.1534/genetics.111.131326. Epub 2011 Aug 11. Genetics. 2011. PMID: 21840861 Free PMC article.
-
Elevated mutation rate during meiosis in Saccharomyces cerevisiae.PLoS Genet. 2015 Jan 8;11(1):e1004910. doi: 10.1371/journal.pgen.1004910. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569256 Free PMC article.
References
-
- Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. - PubMed
-
- Yu HG, Koshland D. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell. 2005;123:397–407. - PubMed
-
- Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. - PubMed
-
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. - PubMed
-
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

