A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo
- PMID: 20865163
- PMCID: PMC2928783
- DOI: 10.1371/journal.ppat.1001064
A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo
Abstract
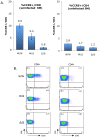
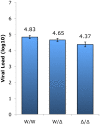
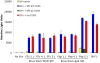
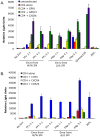
In contrast to HIV infection in humans and SIV in macaques, SIV infection of natural hosts including sooty mangabeys (SM) is non-pathogenic despite robust virus replication. We identified a novel SM CCR5 allele containing a two base pair deletion (Δ2) encoding a truncated molecule that is not expressed on the cell surface and does not support SIV entry in vitro. The allele was present at a 26% frequency in a large SM colony, along with 3% for a CCR5Δ24 deletion allele that also abrogates surface expression. Overall, 8% of animals were homozygous for defective CCR5 alleles and 41% were heterozygous. The mutant allele was also present in wild SM in West Africa. CD8+ and CD4+ T cells displayed a gradient of CCR5 expression across genotype groups, which was highly significant for CD8+ cells. Remarkably, the prevalence of natural SIVsmm infection was not significantly different in animals lacking functional CCR5 compared to heterozygous and homozygous wild-type animals. Furthermore, animals lacking functional CCR5 had robust plasma viral loads, which were only modestly lower than wild-type animals. SIVsmm primary isolates infected both homozygous mutant and wild-type PBMC in a CCR5-independent manner in vitro, and Envs from both CCR5-null and wild-type infected animals used CXCR6, GPR15 and GPR1 in addition to CCR5 in transfected cells. These data clearly indicate that SIVsmm relies on CCR5-independent entry pathways in SM that are homozygous for defective CCR5 alleles and, while the extent of alternative coreceptor use in SM with CCR5 wild type alleles is uncertain, strongly suggest that SIVsmm tropism and host cell targeting in vivo is defined by the distribution and use of alternative entry pathways in addition to CCR5. SIVsmm entry through alternative pathways in vivo raises the possibility of novel CCR5-negative target cells that may be more expendable than CCR5+ cells and enable the virus to replicate efficiently without causing disease in the face of extremely restricted CCR5 expression seen in SM and several other natural host species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. - PubMed
-
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. - PubMed
-
- Peeters M, Janssens W, Fransen K, Brandful J, Heyndrickx L, et al. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Cote d'Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res Hum Retroviruses. 1994;10:1289–1294. - PubMed
-
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01-AI76174/AI/NIAID NIH HHS/United States
- T32 AI007632/AI/NIAID NIH HHS/United States
- P01 AI076174/AI/NIAID NIH HHS/United States
- R37 AI050529/AI/NIAID NIH HHS/United States
- R01-AI66998/AI/NIAID NIH HHS/United States
- R01 AI058706/AI/NIAID NIH HHS/United States
- R01-AI065325/AI/NIAID NIH HHS/United States
- R01 AI065325/AI/NIAID NIH HHS/United States
- RR-P51-00165/RR/NCRR NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- R37 AI066998/AI/NIAID NIH HHS/United States
- R01-AI035502/AI/NIAID NIH HHS/United States
- R01 AI035502/AI/NIAID NIH HHS/United States
- R01 AI066998/AI/NIAID NIH HHS/United States
- RR-P51-000164/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

