Distinct merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with merkel cell carcinoma
- PMID: 20865165
- PMCID: PMC2928786
- DOI: 10.1371/journal.ppat.1001076
Distinct merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with merkel cell carcinoma
Abstract
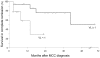
Merkel Cell Polyomavirus (MCPyV) is associated with Merkel Cell carcinoma (MCC), a rare, aggressive skin cancer with neuroendocrine features. The causal role of MCPyV is highly suggested by monoclonal integration of its genome and expression of the viral large T (LT) antigen in MCC cells. We investigated and characterized MCPyV molecular features in MCC, respiratory, urine and blood samples from 33 patients by quantitative PCR, sequencing and detection of integrated viral DNA. We examined associations between either MCPyV viral load in primary MCC or MCPyV DNAemia and survival. Results were interpreted with respect to the viral molecular signature in each compartment. Patients with MCC containing more than 1 viral genome copy per cell had a longer period in complete remission than patients with less than 1 copy per cell (34 vs 10 months, P = 0.037). Peripheral blood mononuclear cells (PBMC) contained MCPyV more frequently in patients sampled with disease than in patients in complete remission (60% vs 11%, P = 0.00083). Moreover, the detection of MCPyV in at least one PBMC sample during follow-up was associated with a shorter overall survival (P = 0.003). Sequencing of viral DNA from MCC and non MCC samples characterized common single nucleotide polymorphisms defining 8 patient specific strains. However, specific molecular signatures truncating MCPyV LT were observed in 8/12 MCC cases but not in respiratory and urinary samples from 15 patients. New integration sites were identified in 4 MCC cases. Finally, mutated-integrated forms of MCPyV were detected in PBMC of two patients with disseminated MCC disease, indicating circulation of metastatic cells. We conclude that MCPyV molecular features in primary MCC tumour and PBMC may help to predict the course of the disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis. 2009;199:837–846. - PubMed
-
- Sharp CP, Norja P, Anthony I, Bell JE, Simmonds P. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. J Infect Dis. 2009;199:398–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

