Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response
- PMID: 20865166
- PMCID: PMC2928790
- DOI: 10.1371/journal.ppat.1001065
Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response
Abstract
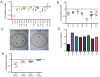
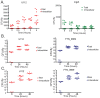
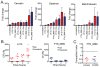
The survival of a bacterial pathogen within a host depends upon its ability to outmaneuver the host immune response. Thus, mutant pathogens provide a useful tool for dissecting host-pathogen relationships, as the strategies the microbe has evolved to counteract immunity reveal a host's immune mechanisms. In this study, we examined the pathogen Francisella novicida and identified new bacterial virulence factors that interact with different parts of the Drosophila melanogaster innate immune system. We performed a genome-wide screen to identify F. novicida genes required for growth and survival within the fly and identified a set of 149 negatively selected mutants. Among these, we identified a class of genes including the transcription factor oxyR, and the DNA repair proteins uvrB, recB, and ruvC that help F. novicida resist oxidative stress. We determined that these bacterial genes are virulence factors that allow F. novicida to counteract the fly melanization immune response. We then performed a second in vivo screen to identify an additional subset of bacterial genes that interact specifically with the imd signaling pathway. Most of these mutants have decreased resistance to the antimicrobial peptide polymyxin B. Characterization of a mutation in the putative transglutaminase FTN_0869 produced a curious result that could not easily be explained using known Drosophila immune responses. By using an unbiased genetic screen, these studies provide a new view of the Drosophila immune response from the perspective of a pathogen. We show that two branches of the fly's immunity are important for fighting F. novicida infections in a model host: melanization and an imd-regulated immune response, and identify bacterial genes that specifically counteract these host responses. Our work suggests that there may be more to learn about the fly immune system, as not all of the phenotypes we observe can be readily explained by its interactions with known immune responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Persson J, Vance RE. Genetics-squared: combining host and pathogen genetics in the analysis of innate immunity and bacterial virulence. Immunogenetics. 2007;59:761–778. - PubMed
-
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. - PubMed
-
- Rosetto M, Engstrom Y, Baldari CT, Telford JL, Hultmark D. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem Biophys Res Commun. 1995;209:111–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

