The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis
- PMID: 20865802
- PMCID: PMC4355639
- DOI: 10.1038/ncomms1082
The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis
Abstract
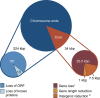
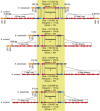
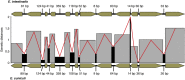
The genome of the microsporidia Encephalitozoon cuniculi is widely recognized as a model for extreme reduction and compaction. At only 2.9 Mbp, the genome encodes approximately 2,000 densely packed genes and little else. However, the nuclear genome of its sister, Encephalitozoon intestinalis, is even more reduced; at 2.3 Mbp, it represents a 20% reduction from an already severely compacted genome, raising the question, what else can be lost? In this paper, we describe the complete sequence of the E. intestinalis genome and its comparison with that of E. cuniculi. The two species share a conserved gene content, order and density over most of their genomes. The exceptions are the subtelomeric regions, where E. intestinalis chromosomes are missing large gene blocks of sequence found in E. cuniculi. In the remaining gene-dense chromosome 'cores', the diminutive intergenic sequences and introns are actually more highly conserved than the genes themselves, suggesting that they have reached the limits of reduction for a fully functional genome.
Figures

References
-
- Corradi N. & Keeling P. J. Microsporidia: a journey through radical taxonomical revisions. Fungal Biol. Rev. 23, 1–8 (2009).
-
- James T. Y. et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822 (2006). - PubMed
-
- Katinka M. D. et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453 (2001). - PubMed
-
- Van de Peer Y., Ben Ali A. & Meyer A. Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene 246, 1–8 (2000). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

