Structure and mechanism of ORF36, an amino sugar oxidizing enzyme in everninomicin biosynthesis
- PMID: 20866105
- PMCID: PMC2964426
- DOI: 10.1021/bi101336u
Structure and mechanism of ORF36, an amino sugar oxidizing enzyme in everninomicin biosynthesis
Abstract
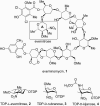
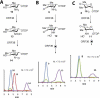
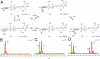
Everninomicin is a highly modified octasaccharide that belongs to the orthosomycin family of antibiotics and possesses potent Gram-positive antibiotic activity, including broad-spectrum efficacy against multidrug resistant enterococci and Staphylococcus aureus. Among its distinctive structural features is a nitro sugar, l-evernitrose, analogues of which decorate a variety of natural products. Recently, we identified a nitrososynthase enzyme encoded by orf36 from Micromonospora carbonacea var. africana that mediates the flavin-dependent double oxidation of synthetically generated thymidine diphosphate (TDP)-l-epi-vancosamine to the corresponding nitroso sugar. Herein, we utilize a five-enzyme in vitro pathway both to verify that ORF36 catalyzes oxidation of biogenic TDP-l-epi-vancosamine and to determine whether ORF36 exhibits catalytic competence for any of its biosynthetic progenitors, which are candidate substrates for nitrososynthases in vivo. Progenitors solely undergo single-oxidation reactions and terminate in the hydroxylamine oxidation state. Performing the in vitro reactions in the presence of (18)O(2) establishes that molecular oxygen, rather than oxygen from water, is incorporated into ORF36-generated intermediates and products and identifies an off-pathway product that correlates with the oxidation product of a progenitor substrate. The 3.15 Å resolution X-ray crystal structure of ORF36 reveals a tetrameric enzyme that shares a fold with acyl-CoA dehydrogenases and class D flavin-containing monooxygenases, including the nitrososynthase KijD3. However, ORF36 and KijD3 have unusually open active sites in comparison to these related enzymes. Taken together, these studies map substrate determinants and allow the proposal of a minimal monooxygenase mechanism for amino sugar oxidation by ORF36.
Figures
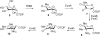

Similar articles
-
MD and QM/MM study on catalytic mechanism of a FAD-dependent enzyme ORF36: for nitro sugar biosynthesis.J Mol Graph Model. 2013 Jul;44:9-16. doi: 10.1016/j.jmgm.2013.04.008. Epub 2013 May 14. J Mol Graph Model. 2013. PMID: 23735899
-
Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):11547-52. doi: 10.1073/pnas.1500964112. Epub 2015 Aug 3. Proc Natl Acad Sci U S A. 2015. PMID: 26240321 Free PMC article.
-
The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.Biochemistry. 2018 Dec 18;57(50):6827-6837. doi: 10.1021/acs.biochem.8b00836. Epub 2018 Dec 7. Biochemistry. 2018. PMID: 30525509 Free PMC article.
-
Diiron monooxygenases in natural product biosynthesis.Nat Prod Rep. 2018 Jul 18;35(7):646-659. doi: 10.1039/C7NP00061H. Nat Prod Rep. 2018. PMID: 29552683 Free PMC article. Review.
-
Pharmacologic and bacteriologic properties of SCH-27899 (Ziracin), an investigational antibiotic from the everninomicin family.Pharmacotherapy. 1999 Oct;19(10):1111-7. doi: 10.1592/phco.19.15.1111.30576. Pharmacotherapy. 1999. PMID: 10512059 Review.
Cited by
-
Bifunctional Nitrone-Conjugated Secondary Metabolite Targeting the Ribosome.J Am Chem Soc. 2020 Oct 28;142(43):18369-18377. doi: 10.1021/jacs.0c04675. Epub 2020 Oct 19. J Am Chem Soc. 2020. PMID: 32709196 Free PMC article.
-
Initial investigations of C4a-(hydro)peroxyflavin intermediate formation by dibenzothiophene monooxygenase.Biochem Biophys Res Commun. 2016 Dec 2;481(1-2):189-194. doi: 10.1016/j.bbrc.2016.10.145. Epub 2016 Nov 1. Biochem Biophys Res Commun. 2016. PMID: 27815073 Free PMC article.
-
Stabilization of C4a-hydroperoxyflavin in a two-component flavin-dependent monooxygenase is achieved through interactions at flavin N5 and C4a atoms.J Biol Chem. 2011 Aug 12;286(32):28170-80. doi: 10.1074/jbc.M111.241836. Epub 2011 Jun 16. J Biol Chem. 2011. PMID: 21680741 Free PMC article.
-
Nitrososynthase-triggered oxidative carbon-carbon bond cleavage in baumycin biosynthesis.J Am Chem Soc. 2013 Aug 7;135(31):11457-60. doi: 10.1021/ja404987r. Epub 2013 Jul 25. J Am Chem Soc. 2013. PMID: 23885759 Free PMC article.
-
Structural basis of arrestin-3 activation and signaling.Nat Commun. 2017 Nov 10;8(1):1427. doi: 10.1038/s41467-017-01218-8. Nat Commun. 2017. PMID: 29127291 Free PMC article.
References
-
- Ganguly AK. Ziracin, a novel oligosaccharide antibiotic. J. Antibiot. 2000;53:1038–1044. - PubMed
-
- Jones RN, Hare RS, Sabatelli FJ. In vitro Gram-positive antimicrobial activity of evernimicin (SCH 27899), a novel oligosaccharide, compared with other antimicrobials: a multicentre international trial. J. Antimicrob. Chemother. 2001;47:15–25. - PubMed
-
- Chu M, Mierzwa R, Jenkins J, Chan TM, Das P, Pramanik B, Patel M, Gullo V. Isolation and characterization of novel oligosaccharides related to Ziracin. J. Nat. Prod. 2002;65:1588–1593. - PubMed
-
- McNicholas PM, Najarian DJ, Mann PA, Hesk D, Hare RS, Shaw KJ, Black TA. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob. Agents. Chemother. 2000;44:1121–1126. - PMC - PubMed
-
- Waitz JA, Horan AC. Micromonospora carbonacea var. africana. Schering Corporation; U.S.: 1988.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

