Subdiffusive motion of a polymer composed of subdiffusive monomers
- PMID: 20866654
- PMCID: PMC4918243
- DOI: 10.1103/PhysRevE.82.011913
Subdiffusive motion of a polymer composed of subdiffusive monomers
Abstract
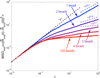
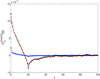
We use Brownian dynamics simulations and analytical theory to investigate the physical principles underlying subdiffusive motion of a polymer. Specifically, we examine the consequences of confinement, self-interaction, viscoelasticity, and random waiting on monomer motion, as these physical phenomena may be relevant to the behavior of biological macromolecules in vivo. We find that neither confinement nor self-interaction alter the fundamental Rouse mode relaxations of a polymer. However, viscoelasticity, modeled by fractional Langevin motion, and random waiting, modeled with a continuous time random walk, lead to significant and distinct deviations from the classic polymer-dynamics model. Our results provide diagnostic tools--the monomer mean square displacement scaling and the velocity autocorrelation function--that can be applied to experimental data to determine the underlying mechanism for subdiffusive motion of a polymer.
Figures


Similar articles
-
Meaningful interpretation of subdiffusive measurements in living cells (crowded environment) by fluorescence fluctuation microscopy.Curr Pharm Biotechnol. 2010 Aug;11(5):527-43. doi: 10.2174/138920110791591454. Curr Pharm Biotechnol. 2010. PMID: 20553227 Free PMC article.
-
Rouse dynamics of colloids bound to polymer networks.Phys Rev Lett. 2007 Nov 16;99(20):208301. doi: 10.1103/PhysRevLett.99.208301. Epub 2007 Nov 15. Phys Rev Lett. 2007. PMID: 18233191
-
Fractional Brownian motion and the critical dynamics of zipping polymers.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Mar;85(3 Pt 1):031120. doi: 10.1103/PhysRevE.85.031120. Epub 2012 Mar 16. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 22587051
-
Single-polymer dynamics under constraints: scaling theory and computer experiment.J Phys Condens Matter. 2011 Mar 16;23(10):103101. doi: 10.1088/0953-8984/23/10/103101. Epub 2011 Feb 18. J Phys Condens Matter. 2011. PMID: 21335636 Review.
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
Cited by
-
How Transcription Factor Clusters Shape the Transcriptional Landscape.Biomolecules. 2024 Jul 20;14(7):875. doi: 10.3390/biom14070875. Biomolecules. 2024. PMID: 39062589 Free PMC article. Review.
-
Fluctuating Diffusivity of RNA-Protein Particles: Analogy with Thermodynamics.Entropy (Basel). 2021 Mar 12;23(3):333. doi: 10.3390/e23030333. Entropy (Basel). 2021. PMID: 33808996 Free PMC article.
-
Probing local chromatin dynamics by tracking telomeres.Biophys J. 2022 Jul 19;121(14):2684-2692. doi: 10.1016/j.bpj.2022.06.020. Epub 2022 Jun 22. Biophys J. 2022. PMID: 35733342 Free PMC article.
-
Dynamics of chromosome organization in a minimal bacterial cell.Front Cell Dev Biol. 2023 Aug 9;11:1214962. doi: 10.3389/fcell.2023.1214962. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37621774 Free PMC article.
-
Escherichia coli Chromosomal Loci Segregate from Midcell with Universal Dynamics.Biophys J. 2016 Jun 21;110(12):2597-2609. doi: 10.1016/j.bpj.2016.04.046. Biophys J. 2016. PMID: 27332118 Free PMC article.
References
-
- Golding I, Cox EC. Phys Rev Lett. 2006;96:098102. - PubMed
-
- Tolic-Nørrelykke IM, Munteanu E-L, Thon G, Oddershede L, Berg-Sørensen K. Phys Rev Lett. 2004;93:078102. - PubMed
-
- Gittes F, Schnurr B, Olmsted P, MacKintosh F, Schmidt C. Phys Rev Lett. 1997;79:3286.
-
- Bouchaud JP, Georges A. Phys Rep. 1990;195:127.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
