HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation
- PMID: 20870715
- PMCID: PMC2988339
- DOI: 10.1074/jbc.M110.133181
HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation
Abstract
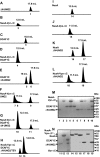
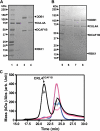
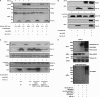
The human immunodeficiency virus type 1 (HIV-1) accessory protein, Vpr, interacts with several host cellular proteins including uracil DNA glycosylase-2 (UNG2) and a cullin-RING E3 ubiquitin ligase assembly (CRL4(DCAF1)). The ligase is composed of cullin 4A (CUL4A), RING H2 finger protein (RBX1), DNA damage-binding protein 1 (DDB1), and a substrate recognition subunit, DDB1- and CUL4-associated factor 1 (DCAF1). Here we show that recombinant UNG2 specifically interacts with Vpr, but not with Vpx of simian immunodeficiency virus, forming a heterotrimeric complex with DCAF1 and Vpr in vitro as well as in vivo. Using reconstituted CRL4(DCAF1) and CRL4(DCAF1-Vpr) E3 ubiquitin ligases in vitro reveals that UNG2 ubiquitination (ubiquitylation) is facilitated by Vpr. Co-expression of DCAF1 and Vpr causes down-regulation of UNG2 in a proteasome-dependent manner, with Vpr mutants that are defective in UNG2 or DCAF1 binding abrogating this effect. Taken together, our results show that the CRL4(DCAF1) E3 ubiquitin ligase can be subverted by Vpr to target UNG2 for degradation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

