The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression
- PMID: 20870725
- PMCID: PMC2988377
- DOI: 10.1074/jbc.M110.139527
The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression
Abstract
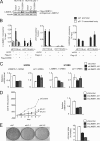
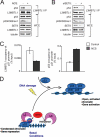
The p53 tumor suppressor protein is regulated by multiple post-translational modifications, including lysine methylation. We previously found that monomethylation of p53 at lysine 382 (p53K382me1) by the protein lysine methyltransferase (PKMT) SET8/PR-Set7 represses p53 transactivation of target genes. However, the molecular mechanism linking p53K382 monomethylation to repression is not known. Here we show in biochemical and crystallographic studies the preferential recognition of p53K382me1 by the triple malignant brain tumor (MBT) repeats of the chromatin compaction factor L3MBTL1. We demonstrate that SET8-mediated methylation of p53 at Lys-382 promotes the interaction between L3MBTL1 and p53 in cells, and the chromatin occupancy of L3MBTL1 at p53 target promoters. In the absence of DNA damage, L3MBTL1 interacts with p53K382me1 and p53-target genes are repressed, whereas depletion of L3MBTL1 results in a p53-dependent increase in p21 and PUMA transcript levels. Activation of p53 by DNA damage is coupled to a decrease in p53K382me1 levels, abrogation of the L3MBTL1-p53 interaction, and disassociation of L3MBTL1 from p53-target promoters. Together, we identify L3MBTL1 as the second known methyl-p53 effector protein, and provide a molecular explanation for the mechanism by which p53K382me1 is transduced to regulate p53 activity.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

