A Gibbs sampling strategy applied to the mapping of ambiguous short-sequence tags
- PMID: 20871106
- PMCID: PMC2951085
- DOI: 10.1093/bioinformatics/btq460
A Gibbs sampling strategy applied to the mapping of ambiguous short-sequence tags
Abstract
Motivation: Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) is widely used in biological research. ChIP-seq experiments yield many ambiguous tags that can be mapped with equal probability to multiple genomic sites. Such ambiguous tags are typically eliminated from consideration resulting in a potential loss of important biological information.
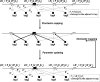
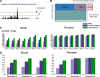
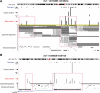
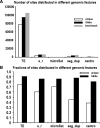
Results: We have developed a Gibbs sampling-based algorithm for the genomic mapping of ambiguous sequence tags. Our algorithm relies on the local genomic tag context to guide the mapping of ambiguous tags. The Gibbs sampling procedure we use simultaneously maps ambiguous tags and updates the probabilities used to infer correct tag map positions. We show that our algorithm is able to correctly map more ambiguous tags than existing mapping methods. Our approach is also able to uncover mapped genomic sites from highly repetitive sequences that can not be detected based on unique tags alone, including transposable elements, segmental duplications and peri-centromeric regions. This mapping approach should prove to be useful for increasing biological knowledge on the too often neglected repetitive genomic regions.
Availability: http://esbg.gatech.edu/jordan/software/map
Contact: king.jordan@biology.gatech.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
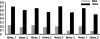
Similar articles
-
piPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing.Bioinformatics. 2015 Feb 15;31(4):593-5. doi: 10.1093/bioinformatics/btu647. Epub 2014 Oct 17. Bioinformatics. 2015. PMID: 25342065 Free PMC article.
-
ArchTEx: accurate extraction and visualization of next-generation sequence data.Bioinformatics. 2012 Apr 1;28(7):1021-3. doi: 10.1093/bioinformatics/bts063. Epub 2012 Feb 2. Bioinformatics. 2012. PMID: 22302569
-
F-Seq: a feature density estimator for high-throughput sequence tags.Bioinformatics. 2008 Nov 1;24(21):2537-8. doi: 10.1093/bioinformatics/btn480. Epub 2008 Sep 10. Bioinformatics. 2008. PMID: 18784119 Free PMC article.
-
Discovering and detecting transposable elements in genome sequences.Brief Bioinform. 2007 Nov;8(6):382-92. doi: 10.1093/bib/bbm048. Epub 2007 Oct 10. Brief Bioinform. 2007. PMID: 17932080 Review.
-
[Processing and analysis of ChIP-seq data].Yi Chuan. 2012 Jun;34(6):773-83. doi: 10.3724/sp.j.1005.2012.00773. Yi Chuan. 2012. PMID: 22698750 Review. Chinese.
Cited by
-
DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape.Nat Genet. 2013 Jul;45(7):836-41. doi: 10.1038/ng.2649. Epub 2013 May 26. Nat Genet. 2013. PMID: 23708189 Free PMC article.
-
Rapid, Paralog-Sensitive CNV Analysis of 2457 Human Genomes Using QuicK-mer2.Genes (Basel). 2020 Jan 29;11(2):141. doi: 10.3390/genes11020141. Genes (Basel). 2020. PMID: 32013076 Free PMC article.
-
On the presence and role of human gene-body DNA methylation.Oncotarget. 2012 Apr;3(4):462-74. doi: 10.18632/oncotarget.497. Oncotarget. 2012. PMID: 22577155 Free PMC article.
-
Mobile genomics: tools and techniques for tackling transposons.Philos Trans R Soc Lond B Biol Sci. 2020 Mar 30;375(1795):20190345. doi: 10.1098/rstb.2019.0345. Epub 2020 Feb 10. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32075565 Free PMC article. Review.
-
MMR: a tool for read multi-mapper resolution.Bioinformatics. 2016 Mar 1;32(5):770-2. doi: 10.1093/bioinformatics/btv624. Epub 2015 Oct 30. Bioinformatics. 2016. PMID: 26519503 Free PMC article.
References
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1–10. - PubMed
-
- Faulkner GJ, et al. A rescue strategy for multimapping short sequence tags refines surveys of transcriptional activity by CAGE. Genomics. 2008;91:281–288. - PubMed
-
- Hashimoto T, et al. Probabilistic resolution of multi-mapping reads in massively parallel sequencing data using MuMRescueLite. Bioinformatics. 2009;25:2613–2614. - PubMed

