Selective inhibition of BET bromodomains
- PMID: 20871596
- PMCID: PMC3010259
- DOI: 10.1038/nature09504
Selective inhibition of BET bromodomains
Abstract
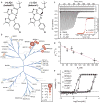
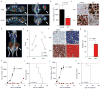
Epigenetic proteins are intently pursued targets in ligand discovery. So far, successful efforts have been limited to chromatin modifying enzymes, or so-called epigenetic 'writers' and 'erasers'. Potent inhibitors of histone binding modules have not yet been described. Here we report a cell-permeable small molecule (JQ1) that binds competitively to acetyl-lysine recognition motifs, or bromodomains. High potency and specificity towards a subset of human bromodomains is explained by co-crystal structures with bromodomain and extra-terminal (BET) family member BRD4, revealing excellent shape complementarity with the acetyl-lysine binding cavity. Recurrent translocation of BRD4 is observed in a genetically-defined, incurable subtype of human squamous carcinoma. Competitive binding by JQ1 displaces the BRD4 fusion oncoprotein from chromatin, prompting squamous differentiation and specific antiproliferative effects in BRD4-dependent cell lines and patient-derived xenograft models. These data establish proof-of-concept for targeting protein-protein interactions of epigenetic 'readers', and provide a versatile chemical scaffold for the development of chemical probes more broadly throughout the bromodomain family.
Figures



Comment in
-
Drug discovery: Reader's block.Nature. 2010 Dec 23;468(7327):1050-1. doi: 10.1038/4681050a. Nature. 2010. PMID: 21179160 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Research Materials

