Structural Optimization of Non-Nucleotide Loop Replacements for Duplex and Triplex DNAs
- PMID: 20871801
- PMCID: PMC2943669
- DOI: 10.1021/ja00126a004
Structural Optimization of Non-Nucleotide Loop Replacements for Duplex and Triplex DNAs
Abstract
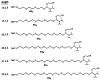
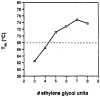
Described are studies systematically exploring structural effects in he use of ethylene glycol (EG) oligomers as non-nucleotide replacements for nucleotide loops in duplex and triplex DNAs. The new structurally optimized loop replacements are more stabilizing in duplexes and triplexes than previously described EG-based linkers. A series of compounds ranging in length from tris(ethylene glycol) to octakis(ethylene glycol) are derivatized as monodimethoxytrityl ethers on one end and phosphoramidites on the other, to enable their incorporation into DNA strands by automated methods. These linker molecules span lengths ranging from 13 to 31 Å in extended conformation. They are incorporated into a series of duplex-forming and triplex-forming sequences, and the stabilities of the corresponding helixes are measured by thermal denaturation. In the duplex series, results show that the optimum linker is the one derived from heptakis(ethylene glycol), which is longer than most previous loop replacements studied. This affords a helix with greater thermal stability than one with a natural T(4) loop. In the triplex series, the loop replacements were examined in four separate situations, in which the loop lies in the 5' or 3' orientation and the central purine target strand is short or extends beyond the loop. Results show that in all cases the loop derived from octakis(ethylene glycol) (EG(8)) gives the greatest stability. In the cases where the target strand is short, the EG(8)-linked probe strands bind with affinities in some cases greater than those with a natural pentanucleotide (T(5)) loop. For the cases where the target strand extends beyond the linker, the EG(8)-linked strands are much lower in the 5' loop orientation than in the 3' loop orientation. It is found that extension by one additional nucleotide in one of the bonding domains in the EG-linked series can result in considerably greater stabilities with long target strands. Overall, the data show that optimum loop replacements are longer than would be expected from simple distance analysis. The results are discussed in relation to expected lengths and geometries for double and triple helixes. The findings will be usefull in the design of synthetically modified nucleic acids for use as diagnostic probes, as biochemical tools, and as potential therapeutic agents.
Figures






Similar articles
-
Flexible non-nucleotide linkers as loop replacements in short double helical RNAs.Nucleic Acids Res. 2000 May 1;28(9):1859-63. doi: 10.1093/nar/28.9.1859. Nucleic Acids Res. 2000. PMID: 10756183 Free PMC article.
-
Targeting pyrimidine single strands by triplex formation: structural optimization of binding.Nucleic Acids Res. 1995 Aug 11;23(15):2937-44. doi: 10.1093/nar/23.15.2937. Nucleic Acids Res. 1995. PMID: 7544889 Free PMC article.
-
Stabilities of double- and triple-strand helical nucleic acids.Prog Biophys Mol Biol. 1992;58(3):225-57. doi: 10.1016/0079-6107(92)90007-s. Prog Biophys Mol Biol. 1992. PMID: 1380719 Review.
-
Evidence for a DNA triplex in a recombination-like motif: I. Recognition of Watson-Crick base pairs by natural bases in a high-stability triplex.J Mol Recognit. 2001 Mar-Apr;14(2):122-39. doi: 10.1002/jmr.528. J Mol Recognit. 2001. PMID: 11301482
-
Triplex DNA structures.Annu Rev Biochem. 1995;64:65-95. doi: 10.1146/annurev.bi.64.070195.000433. Annu Rev Biochem. 1995. PMID: 7574496 Review.
Cited by
-
Stabilization of RNA hairpins using non-nucleotide linkers and circularization.Nucleic Acids Res. 2017 Jun 2;45(10):e92. doi: 10.1093/nar/gkx122. Nucleic Acids Res. 2017. PMID: 28334744 Free PMC article.
-
Recognition of DNA, RNA, and Proteins by Circular Oligonucleotides.Acc Chem Res. 1998 Aug 18;31(8):502-510. doi: 10.1021/ar9602462. Acc Chem Res. 1998. PMID: 19946615 Free PMC article. No abstract available.
-
Efficient production of superior dumbbell-shaped DNA minimal vectors for small hairpin RNA expression.Nucleic Acids Res. 2015 Oct 15;43(18):e120. doi: 10.1093/nar/gkv583. Epub 2015 Jun 11. Nucleic Acids Res. 2015. PMID: 26068470 Free PMC article.
-
Sequence-specific and Selective Recognition of Double-stranded RNAs over Single-stranded RNAs by Chemically Modified Peptide Nucleic Acids.J Vis Exp. 2017 Sep 21;(127):56221. doi: 10.3791/56221. J Vis Exp. 2017. PMID: 28994801 Free PMC article.
-
Loop and backbone modifications of peptide nucleic acid improve g-quadruplex binding selectivity.J Am Chem Soc. 2009 Dec 30;131(51):18415-24. doi: 10.1021/ja907250j. J Am Chem Soc. 2009. PMID: 19947597 Free PMC article.
References
-
- Uhlmann E, Peyman A. Chem. Rev. 1990;90:543.
-
- Beaucage SL, Iyer RP. Tetrahedron. 1993;49:6123–6194.
-
- Giovannangeli C, Montenay-Garestier T, Rougée M, Chassignol M, Thuong NT, Hélène C. J. Am. Chem. Soc. 1991;113:7775–7776.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
