Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression
- PMID: 20874735
- PMCID: PMC2998557
- DOI: 10.1111/j.1558-5646.2010.01136.x
Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression
Abstract
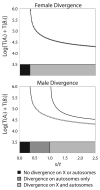
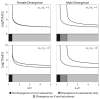
Sex-biased genes--genes that are differentially expressed within males and females--are nonrandomly distributed across animal genomes, with sex chromosomes and autosomes often carrying markedly different concentrations of male- and female-biased genes. These linkage patterns are often gene- and lineage-dependent, differing between functional genetic categories and between species. Although sex-specific selection is often hypothesized to shape the evolution of sex-linked and autosomal gene content, population genetics theory has yet to account for many of the gene- and lineage-specific idiosyncrasies emerging from the empirical literature. With the goal of improving the connection between evolutionary theory and a rapidly growing body of genome-wide empirical studies, we extend previous population genetics theory of sex-specific selection by developing and analyzing a biologically informed model that incorporates sex linkage, pleiotropy, recombination, and epistasis, factors that are likely to vary between genes and between species. Our results demonstrate that sex-specific selection and sex-specific recombination rates can generate, and are compatible with, the gene- and species-specific linkage patterns reported in the genomics literature. The theory suggests that sexual selection may strongly influence the architectures of animal genomes, as well as the chromosomal distribution of fixed substitutions underlying sexually dimorphic traits.
© 2010 The Author(s). Evolution© 2010 The Society for the Study of Evolution.
Figures

References
-
- Agrawal AF. Sexual selection and the maintenance of sexual reproduction. Nature. 2001;411:692–695. - PubMed
-
- Akesson M, Hansson B, Hasselquist D, Bensch S. Linkage mapping of AFLP markers in a wild population of great reed warblers: importance of heterozygosity and number of genotyped individuals. Mol Ecol. 2007;16:2189–2202. - PubMed
-
- Albert AYK, Otto SP. Sexual selection can resolve sex-linked sexual antagonism. Science. 2005;310:119–121. - PubMed
-
- Andersson M. Sexual Selection. Princeton University Press; New Jersey: 1994.
-
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. - PubMed
