Short-hairpin RNA-induced suppression of adenine nucleotide translocase-2 in breast cancer cells restores their susceptibility to TRAIL-induced apoptosis by activating JNK and modulating TRAIL receptor expression
- PMID: 20875141
- PMCID: PMC2955620
- DOI: 10.1186/1476-4598-9-262
Short-hairpin RNA-induced suppression of adenine nucleotide translocase-2 in breast cancer cells restores their susceptibility to TRAIL-induced apoptosis by activating JNK and modulating TRAIL receptor expression
Abstract
Background: Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL; apo2 ligand) induces apoptosis in cancer cells but has little effect on normal cells. However, many cancer cell types are resistant to TRAIL-induced apoptosis, limiting the clinical utility of TRAIL as an anti-cancer agent. We previously reported that the suppression of adenine nucleotide translocase-2 (ANT2) by short-hairpin RNA (shRNA) induces apoptosis of breast cancer cells, which frequently express high levels of ANT2. In the present study, we examined the effect of RNA shRNA-induced suppression of ANT2 on the resistance of breast cancer cells to TRAIL-induced apoptosis in vitro and in vivo.
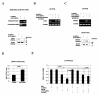
Results: ANT2 shRNA treatment sensitized MCF7, T47 D, and BT474 cells to TRAIL-induced apoptosis by up-regulating the expression of TRAIL death receptors 4 and 5 (DR4 and DR5) and down-regulating the TRAIL decoy receptor 2 (DcR2). In MCF7 cells, ANT2 knockdown activated the stress kinase c-Jun N-terminal kinase (JNK), subsequently stabilizing and increasing the transcriptional activity of p53 by phosphorylating it at Thr81; it also enhanced the expression and activity of DNA methyltransferase 1 (DNMT1). ANT2 shRNA-induced overexpression of DR4/DR5 and TRAIL sensitization were blocked by a p53 inhibitor, suggesting that p53 activation plays an important role in the transcriptional up-regulation of DR4/DR5. However, ANT2 knockdown also up-regulated DR4/DR5 in the p53-mutant cell lines BT474 and T47 D. In MCF7 cells, ANT2 shRNA treatment led to DcR2 promoter methylation and concomitant down-regulation of DcR2 expression, consistent with the observed activation of DNMT1. Treatment of the cells with a demethylating agent or JNK inhibitor prevented the ANT2 shRNA-induced down-regulation of DcR2 and activation of both p53 and DNMT1. In in vivo experiments using nude mice, ANT2 shRNA caused TRAIL-resistant MCF7 xenografts to undergo TRAIL-induced cell death, up-regulated DR4/DR5, and down-regulated DcR2. Co-treatment with ANT2 shRNA and TRAIL efficiently suppressed tumor growth in these mice.
Conclusions: ANT2 suppression by shRNA might be exploited to overcome TRAIL-resistance in cancer.
Figures

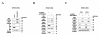



Similar articles
-
Down-regulation of Cbl-b by bufalin results in up-regulation of DR4/DR5 and sensitization of TRAIL-induced apoptosis in breast cancer cells.J Cancer Res Clin Oncol. 2012 Aug;138(8):1279-89. doi: 10.1007/s00432-012-1204-4. Epub 2012 Mar 24. J Cancer Res Clin Oncol. 2012. PMID: 22447040 Free PMC article.
-
Apigenin sensitizes prostate cancer cells to Apo2L/TRAIL by targeting adenine nucleotide translocase-2.PLoS One. 2013;8(2):e55922. doi: 10.1371/journal.pone.0055922. Epub 2013 Feb 19. PLoS One. 2013. PMID: 23431365 Free PMC article.
-
Ursolic acid, a pentacyclin triterpene, potentiates TRAIL-induced apoptosis through p53-independent up-regulation of death receptors: evidence for the role of reactive oxygen species and JNK.J Biol Chem. 2011 Feb 18;286(7):5546-57. doi: 10.1074/jbc.M110.183699. Epub 2010 Dec 14. J Biol Chem. 2011. Retraction in: J Biol Chem. 2016 Aug 5;291(32):16924. doi: 10.1074/jbc.A110.183699. PMID: 21156789 Free PMC article. Retracted.
-
Regulation of TNF-Related Apoptosis-Inducing Ligand Signaling by Glycosylation.Int J Mol Sci. 2018 Mar 2;19(3):715. doi: 10.3390/ijms19030715. Int J Mol Sci. 2018. PMID: 29498673 Free PMC article. Review.
-
TRAIL of Hope Meeting Resistance in Cancer.Trends Cancer. 2020 Dec;6(12):989-1001. doi: 10.1016/j.trecan.2020.06.006. Epub 2020 Jul 24. Trends Cancer. 2020. PMID: 32718904 Free PMC article. Review.
Cited by
-
Cyclophilin 40 alters UVA-induced apoptosis and mitochondrial ROS generation in keratinocytes.Exp Cell Res. 2013 Mar 10;319(5):750-60. doi: 10.1016/j.yexcr.2012.11.016. Epub 2012 Dec 3. Exp Cell Res. 2013. PMID: 23220213 Free PMC article.
-
Gene silencing of FANCF potentiates the sensitivity to mitoxantrone through activation of JNK and p38 signal pathways in breast cancer cells.PLoS One. 2012;7(8):e44254. doi: 10.1371/journal.pone.0044254. Epub 2012 Aug 28. PLoS One. 2012. PMID: 22952942 Free PMC article.
-
ANT2 shRNA downregulates miR-19a and miR-96 through the PI3K/Akt pathway and suppresses tumor growth in hepatocellular carcinoma cells.Exp Mol Med. 2016 Mar 25;48(3):e222. doi: 10.1038/emm.2015.126. Exp Mol Med. 2016. PMID: 27012708 Free PMC article.
-
Synergistic Effect of Subtoxic-dose Cisplatin and TRAIL to Mediate Apoptosis by Down-regulating Decoy Receptor 2 and Up-regulating Caspase-8, Caspase-9 and Bax Expression on NCI-H460 and A549 Cells.Iran J Basic Med Sci. 2013 May;16(5):710-8. Iran J Basic Med Sci. 2013. PMID: 23826494 Free PMC article.
-
Breast cancer proteome takes more than two to tango on TRAIL: beat them at their own game.J Membr Biol. 2012 Dec;245(12):763-77. doi: 10.1007/s00232-012-9490-y. Epub 2012 Aug 17. J Membr Biol. 2012. PMID: 22899350 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

