Hepatitis E virus genotype diversity in eastern China
- PMID: 20875298
- PMCID: PMC3294409
- DOI: 10.3201/eid1610.100873
Hepatitis E virus genotype diversity in eastern China
Abstract
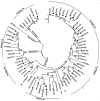
We studied 47 hepatitis E virus (HEV) isolates from hospitalized patients in Nanjing and Taizhou, eastern China. Genotypes 1, 3, and 4 were prevalent; genotype 3 and subgenotype 4b showed a close relationship with the swine strains in eastern China, thus indicating that HEV genotype 3 had infected humans in China.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
