The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration
- PMID: 20875423
- PMCID: PMC3001104
- DOI: 10.1016/j.jmb.2010.09.043
The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration
Abstract
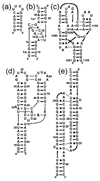
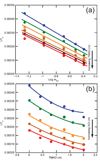
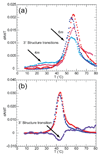
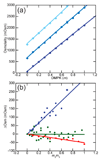
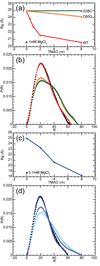
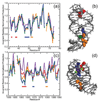
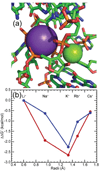
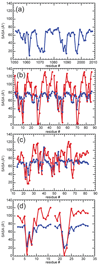
The stabilization of RNA tertiary structures by ions is well known, but the neutral osmolyte trimethylamine oxide (TMAO) can also effectively stabilize RNA tertiary structure. To begin to understand the physical basis for the effects of TMAO on RNA, we have quantitated the TMAO-induced stabilization of five RNAs with known structures. So-called m values, the increment in unfolding free energy per molal of osmolyte at constant KCl activity, are ∼0 for a hairpin secondary structure and between 0.70 and 1.85 kcal mol(-1)m(-1) for four RNA tertiary structures (30-86 nt). Further analysis of two RNAs by small-angle X-ray scattering and hydroxyl radical probing shows that TMAO reduces the radius of gyration of the unfolded ensemble to the same endpoint as seen in titration with Mg(2+) and that the structures stabilized by TMAO and Mg(2+) are indistinguishable. Remarkably, TMAO induces the native conformation of a Mg(2+) ion chelation site formed in part by a buried phosphate, even though Mg(2+) is absent. TMAO interacts weakly, if at all, with KCl, ruling out the possibility that TMAO stabilizes RNA indirectly by increasing salt activity. TMAO is, however, strongly excluded from the vicinity of dimethylphosphate (unfavorable interaction free energy, +211 cal mol(-1)m(-1) for the potassium salt), an ion that mimics the RNA backbone phosphate. We suggest that formation of RNA tertiary structure is accompanied by substantial phosphate dehydration (loss of 66-173 water molecules in the RNA structures studied) and that TMAO works principally by reducing the energetic penalty associated with this dehydration. The strong parallels we find between the effects of TMAO and Mg(2+) suggest that RNA sequence is more important than specific ion interactions in specifying the native structure.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. - PubMed
-
- Liu Y, Bolen DW. The peptide backbone plays a dominant role in protein stabilization by naturally occurring osmolytes. Biochemistry. 1995;34:12884–12891. - PubMed
-
- Timasheff SN. Control of protein stability and reactions by weakly interacting cosolvents: the simplicity of the complicated. Adv Protein Chem. 1998;51:355–432. - PubMed
-
- Courtenay ES, Capp MW, Saecker RM, Record MT., Jr Thermodynamic analysis of interactions between denaturants and protein surface exposed on unfolding: interpretation of urea and guanidinium chloride m-values and their correlation with changes in accessible surface area (ASA) using preferential interaction coefficients and the local-bulk domain model. Proteins Suppl. 2000;4:72–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

