Dynamic interactions between clathrin and locally structured elements in a disordered protein mediate clathrin lattice assembly
- PMID: 20875424
- PMCID: PMC2981644
- DOI: 10.1016/j.jmb.2010.09.044
Dynamic interactions between clathrin and locally structured elements in a disordered protein mediate clathrin lattice assembly
Abstract
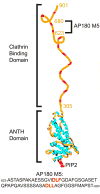
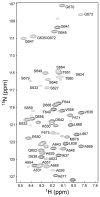
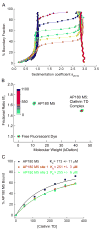
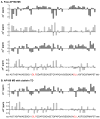
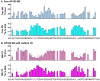
Assembly of clathrin lattices is mediated by assembly/adaptor proteins that contain domains that bind lipids or membrane-bound cargo proteins and clathrin binding domains (CBDs) that recruit clathrin. Here, we characterize the interaction between clathrin and a large fragment of the CBD of the clathrin assembly protein AP180. Mutational, NMR chemical shift, and analytical ultracentrifugation analyses allowed us to precisely define two clathrin binding sites within this fragment, each of which is found to bind weakly to the N-terminal domain of the clathrin heavy chain (TD). The locations of the two clathrin binding sites are consistent with predictions from sequence alignments of previously identified clathrin binding elements and, by extension, indicate that the complete AP180 CBD contains ∼12 degenerate repeats, each containing a single clathrin binding site. Sequence and circular dichroism analyses have indicated that the AP180 CBD is predominantly unstructured and our NMR analyses confirm that this is largely the case for the AP180 fragment characterized here. Unexpectedly, unlike the many proteins that undergo binding-coupled folding upon interaction with their binding partners, the AP180 fragment is similarly unstructured in its bound and free states. Instead, we find that this fragment exhibits localized β-turn-like structures at the two clathrin binding sites both when free and when bound to clathrin. These observations are incorporated into a model in which weak binding by multiple, pre-structured clathrin binding elements regularly dispersed throughout a largely unstructured CBD allows efficient recruitment of clathrin to endocytic sites and dynamic assembly of the clathrin lattice.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


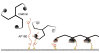
Similar articles
-
Molecular and functional characterization of clathrin- and AP-2-binding determinants within a disordered domain of auxilin.J Biol Chem. 2003 Jul 11;278(28):25357-68. doi: 10.1074/jbc.M303738200. Epub 2003 May 5. J Biol Chem. 2003. PMID: 12732633
-
Identification and functional characterization of Arabidopsis AP180, a binding partner of plant alphaC-adaptin.J Cell Sci. 2004 Apr 15;117(Pt 10):2051-62. doi: 10.1242/jcs.01062. Epub 2004 Mar 30. J Cell Sci. 2004. PMID: 15054111
-
A Novel Sequence in AP180 and CALM Promotes Efficient Clathrin Binding and Assembly.PLoS One. 2016 Aug 30;11(8):e0162050. doi: 10.1371/journal.pone.0162050. eCollection 2016. PLoS One. 2016. PMID: 27574975 Free PMC article.
-
Getting in touch with the clathrin terminal domain.Traffic. 2012 Apr;13(4):511-9. doi: 10.1111/j.1600-0854.2011.01321.x. Epub 2012 Jan 13. Traffic. 2012. PMID: 22239657 Free PMC article. Review.
-
AP180 N-Terminal Homology (ANTH) and Epsin N-Terminal Homology (ENTH) Domains: Physiological Functions and Involvement in Disease.Adv Exp Med Biol. 2019;1111:55-76. doi: 10.1007/5584_2018_218. Adv Exp Med Biol. 2019. PMID: 29774507 Review.
Cited by
-
Nuclear Magnetic Resonance Structural Mapping Reveals Promiscuous Interactions between Clathrin-Box Motif Sequences and the N-Terminal Domain of the Clathrin Heavy Chain.Biochemistry. 2015 Apr 28;54(16):2571-80. doi: 10.1021/acs.biochem.5b00065. Epub 2015 Apr 16. Biochemistry. 2015. PMID: 25844500 Free PMC article.
-
Molecular Mechanisms of Membrane Curvature Sensing by a Disordered Protein.J Am Chem Soc. 2019 Jul 3;141(26):10361-10371. doi: 10.1021/jacs.9b03927. Epub 2019 Jun 20. J Am Chem Soc. 2019. PMID: 31180661 Free PMC article.
-
The ∼ 16 kDa C-terminal sequence of clathrin assembly protein AP180 is essential for efficient clathrin binding.PLoS One. 2014 Oct 20;9(10):e110557. doi: 10.1371/journal.pone.0110557. eCollection 2014. PLoS One. 2014. PMID: 25329427 Free PMC article.
-
A conserved interaction between the effector Sca4 and host clathrin suggests additional contributions for Sca4 during rickettsial infection.Infect Immun. 2024 Dec 10;92(12):e0026724. doi: 10.1128/iai.00267-24. Epub 2024 Nov 13. Infect Immun. 2024. PMID: 39535192 Free PMC article.
-
Liquid-like protein interactions catalyse assembly of endocytic vesicles.Nat Cell Biol. 2021 Apr;23(4):366-376. doi: 10.1038/s41556-021-00646-5. Epub 2021 Apr 5. Nat Cell Biol. 2021. PMID: 33820972 Free PMC article.
References
-
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. - PubMed
-
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–7. - PubMed
-
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–96. - PubMed
-
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–5. - PubMed
-
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

