Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action
- PMID: 20876128
- PMCID: PMC2951456
- DOI: 10.1073/pnas.1007988107
Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action
Abstract
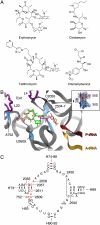
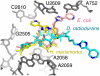
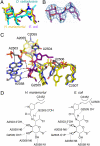
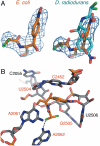
Differences between the structures of bacterial, archaeal, and eukaryotic ribosomes account for the selective action of antibiotics. Even minor variations in the structure of ribosomes of different bacterial species may lead to idiosyncratic, species-specific interactions of the drugs with their targets. Although crystallographic structures of antibiotics bound to the peptidyl transferase center or the exit tunnel of archaeal (Haloarcula marismortui) and bacterial (Deinococcus radiodurans) large ribosomal subunits have been reported, it remains unclear whether the interactions of antibiotics with these ribosomes accurately reflect those with the ribosomes of pathogenic bacteria. Here we report X-ray crystal structures of the Escherichia coli ribosome in complexes with clinically important antibiotics of four major classes, including the macrolide erythromycin, the ketolide telithromycin, the lincosamide clindamycin, and a phenicol, chloramphenicol, at resolutions of ∼3.3 Å-3.4 Å. Binding modes of three of these antibiotics show important variations compared to the previously determined structures. Biochemical and structural evidence also indicates that interactions of telithromycin with the E. coli ribosome more closely resembles drug binding to ribosomes of bacterial pathogens. The present data further argue that the identity of nucleotides 752, 2609, and 2055 of 23S ribosomal RNA explain in part the spectrum and selectivity of antibiotic action.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Designer drugs for discerning bugs.Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17065-6. doi: 10.1073/pnas.1012547107. Epub 2010 Sep 27. Proc Natl Acad Sci U S A. 2010. PMID: 20876111 Free PMC article. No abstract available.
References
-
- Cundcliffe E. Antibiotic biosynthesis: Some thoughts on “why” and “how”. In: Garrett RA, Liljas A, Matheson AT, Moore PB, Noller HF, editors. The Ribosome:Structure, Function, Antibiotics and Cellular Interactions. Washington, DC: ASM Press; 2000. pp. 409–417.
-
- Sohmen D, Harms JM, Schluenzen F, Wilson DN. SnapShot: Antibiotic inhibition of protein synthesis II. Cell. 2009;139 212.e211. - PubMed
-
- Polacek N, Mankin AS. The ribosomal peptidyl transferase center: Structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol. 2005;40:285–311. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

