Revisiting the structures of several antibiotics bound to the bacterial ribosome
- PMID: 20876130
- PMCID: PMC2951403
- DOI: 10.1073/pnas.1008685107
Revisiting the structures of several antibiotics bound to the bacterial ribosome
Abstract
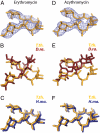
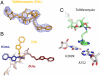
The increasing prevalence of antibiotic-resistant pathogens reinforces the need for structures of antibiotic-ribosome complexes that are accurate enough to enable the rational design of novel ribosome-targeting therapeutics. Structures of many antibiotics in complex with both archaeal and eubacterial ribosomes have been determined, yet discrepancies between several of these models have raised the question of whether these differences arise from species-specific variations or from experimental problems. Our structure of chloramphenicol in complex with the 70S ribosome from Thermus thermophilus suggests a model for chloramphenicol bound to the large subunit of the bacterial ribosome that is radically different from the prevailing model. Further, our structures of the macrolide antibiotics erythromycin and azithromycin in complex with a bacterial ribosome are indistinguishable from those determined of complexes with the 50S subunit of Haloarcula marismortui, but differ significantly from the models that have been published for 50S subunit complexes of the eubacterium Deinococcus radiodurans. Our structure of the antibiotic telithromycin bound to the T. thermophilus ribosome reveals a lactone ring with a conformation similar to that observed in the H. marismortui and D. radiodurans complexes. However, the alkyl-aryl moiety is oriented differently in all three organisms, and the contacts observed with the T. thermophilus ribosome are consistent with biochemical studies performed on the Escherichia coli ribosome. Thus, our results support a mode of macrolide binding that is largely conserved across species, suggesting that the quality and interpretation of electron density, rather than species specificity, may be responsible for many of the discrepancies between the models.
Conflict of interest statement
Conflict of interest statement: Thomas A. Steitz is a scientific advisor for Rib-X Pharmaceuticals, a product-driven small molecule drug discovery and development company focused on the structure-based design of unique classes of antibiotics.
Figures
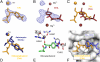
Comment in
-
Designer drugs for discerning bugs.Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17065-6. doi: 10.1073/pnas.1012547107. Epub 2010 Sep 27. Proc Natl Acad Sci U S A. 2010. PMID: 20876111 Free PMC article. No abstract available.
References
-
- Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. - PubMed
-
- Ardanuy C, et al. Molecular characterization of macrolide- and multidrug-resistant Streptococcus pyogenes isolated from adult patients in Barcelona, Spain (1993–2008) J Antimicrob Chemother. 2010;65:634–643. - PubMed
-
- Takaya A, et al. Mutational analysis of reduced telithromycin susceptibility of Streptococcus pneumoniae isolated clinically in Japan. FEMS Microbiol Lett. 2010;307:87–93. - PubMed
-
- Morozumi M, et al. Macrolide-resistant Mycoplasma pneumoniae: Characteristics of isolates and clinical aspects of community-acquired pneumonia. J Infect Chemother. 2010;16:78–86. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

