Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis
- PMID: 20876132
- PMCID: PMC2955109
- DOI: 10.1073/pnas.1006848107
Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis
Abstract
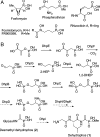
Phosphonate natural products possess a range of biological activities as a consequence of their ability to mimic phosphate esters or tetrahedral intermediates formed in enzymatic reactions involved in carboxyl group metabolism. The dianionic form of these compounds at pH 7 poses a drawback with respect to their ability to mimic carboxylates and tetrahedral intermediates. Microorganisms producing phosphonates have evolved two solutions to overcome this hurdle: biosynthesis of monoanionic phosphinates containing two P-C bonds or esterification of the phosphonate group. The latter solution was first discovered for the antibiotic dehydrophos that contains a methyl ester of a phosphonodehydroalanine group. We report here the expression, purification, substrate scope, and structure of the O-methyltransferase from the dehydrophos biosynthetic gene cluster. The enzyme utilizes S-adenosylmethionine to methylate a variety of phosphonates including 1-hydroxyethylphosphonate, 1,2-dihydroxyethylphosphonate, and acetyl-1-aminoethylphosphonate. Kinetic analysis showed that the best substrates are tripeptides containing as C-terminal residue a phosphonate analog of alanine suggesting the enzyme acts late in the biosynthesis of dehydrophos. These conclusions are corroborated by the X-ray structure that reveals an active site that can accommodate a tripeptide substrate. Furthermore, the structural studies demonstrate a conformational change brought about by substrate or product binding. Interestingly, the enzyme has low substrate specificity and was used to methylate the clinical antibiotic fosfomycin and the antimalaria clinical candidate fosmidomycin, showing its promise for applications in bioengineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
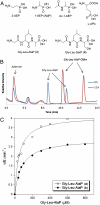
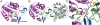
 structure. The sulfate anion is shown in orange, residues that contact the sulfate are shown in yellow, and the intersubunit interaction between Tyr29 and Glu155 is shown in yellow and white, respectively.
structure. The sulfate anion is shown in orange, residues that contact the sulfate are shown in yellow, and the intersubunit interaction between Tyr29 and Glu155 is shown in yellow and white, respectively.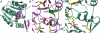
 structure are shown in pink. (B) Close-up view of the composite active site in the DhpI-SAM-
structure are shown in pink. (B) Close-up view of the composite active site in the DhpI-SAM- structure showing the interaction between two monomers, colored in pink and green. Active site residues from the pink monomer are shown in yellow and those from the capping helix from the green monomer are shown in green. (C) A close-up view of the equivalent region in the DhpI-SAH structure, showing the reorganization of the active site residues Tyr29 and Arg168.
structure showing the interaction between two monomers, colored in pink and green. Active site residues from the pink monomer are shown in yellow and those from the capping helix from the green monomer are shown in green. (C) A close-up view of the equivalent region in the DhpI-SAH structure, showing the reorganization of the active site residues Tyr29 and Arg168.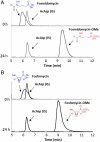
Similar articles
-
Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos, and phosphinothricin.Arch Biochem Biophys. 2011 Jan 1;505(1):13-21. doi: 10.1016/j.abb.2010.09.012. Epub 2010 Sep 18. Arch Biochem Biophys. 2011. PMID: 20854789 Free PMC article. Review.
-
Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology.Curr Opin Chem Biol. 2013 Aug;17(4):580-8. doi: 10.1016/j.cbpa.2013.06.018. Epub 2013 Jul 17. Curr Opin Chem Biol. 2013. PMID: 23870698 Free PMC article. Review.
-
Methylcobalamin-Dependent Radical SAM C-Methyltransferase Fom3 Recognizes Cytidylyl-2-hydroxyethylphosphonate and Catalyzes the Nonstereoselective C-Methylation in Fosfomycin Biosynthesis.Biochemistry. 2017 Jul 18;56(28):3519-3522. doi: 10.1021/acs.biochem.7b00472. Epub 2017 Jul 7. Biochemistry. 2017. PMID: 28678474
-
Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster.Angew Chem Int Ed Engl. 2014 Jan 27;53(5):1334-7. doi: 10.1002/anie.201308363. Epub 2013 Dec 27. Angew Chem Int Ed Engl. 2014. PMID: 24376039 Free PMC article.
-
Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways.J Biol Chem. 2008 Aug 22;283(34):23161-8. doi: 10.1074/jbc.M801788200. Epub 2008 Jun 10. J Biol Chem. 2008. PMID: 18544530 Free PMC article.
Cited by
-
Molecular mechanism of S-adenosylmethionine sensing by SAMTOR in mTORC1 signaling.Sci Adv. 2022 Jul;8(26):eabn3868. doi: 10.1126/sciadv.abn3868. Epub 2022 Jul 1. Sci Adv. 2022. PMID: 35776786 Free PMC article.
-
Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos, and phosphinothricin.Arch Biochem Biophys. 2011 Jan 1;505(1):13-21. doi: 10.1016/j.abb.2010.09.012. Epub 2010 Sep 18. Arch Biochem Biophys. 2011. PMID: 20854789 Free PMC article. Review.
-
Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology.Curr Opin Chem Biol. 2013 Aug;17(4):580-8. doi: 10.1016/j.cbpa.2013.06.018. Epub 2013 Jul 17. Curr Opin Chem Biol. 2013. PMID: 23870698 Free PMC article. Review.
-
Molecular Basis for Autocatalytic Backbone N-Methylation in RiPP Natural Product Biosynthesis.ACS Chem Biol. 2018 Oct 19;13(10):2989-2999. doi: 10.1021/acschembio.8b00668. Epub 2018 Sep 25. ACS Chem Biol. 2018. PMID: 30204409 Free PMC article.
-
Identification and characterization of a welwitindolinone alkaloid biosynthetic gene cluster in the stigonematalean Cyanobacterium Hapalosiphon welwitschii.Chembiochem. 2014 Mar 21;15(5):665-9. doi: 10.1002/cbic.201300794. Chembiochem. 2014. PMID: 24677572 Free PMC article.
References
-
- Seto H, Kuzuyama T. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat Prod Rep. 1999;16:589–596. - PubMed
-
- Woodyer RD, Li G, Zhao H, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem Commun. 2007:359–361. - PubMed
-
- Woodyer RD, et al. Heterologous production of fosfomycin and identification of the minimal biosynthetic cluster. Chem Biol. 2006;13:1171–1182. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

