Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
- PMID: 20876133
- PMCID: PMC2955107
- DOI: 10.1073/pnas.1005681107
Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Abstract
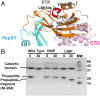
Thermolysin-like proteases (TLPs), a large group of zinc metalloproteases, are synthesized as inactive precursors. TLPs with a long propeptide (∼200 residues) undergo maturation following autoprocessing through an elusive molecular mechanism. We report the first two crystal structures for the autoprocessed complexes of a typical TLP, MCP-02. In the autoprocessed complex, Ala205 shifts upward by 33 Å from the previously covalently linked residue, His204, indicating that, following autocleavage of the peptide bond between His204 and Ala205, a large conformational change from the zymogen to the autoprocessed complex occurs. The eight N-terminal residues (residues Ala205-Gly212) of the catalytic domain form a new β-strand, nestling into two other β-strands. Simultaneously, the apparent T(m) of the autoprocessed complex increases 20 °C compared to that of the zymogen. The stepwise degradation of the propeptide begins with two sequential cuttings at Ser49-Val50 and Gly57-Leu58, which lead to the disassembly of the propeptide and the formation of mature MCP-02. Our findings give new insights into the molecular mechanism of TLP maturation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

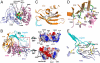


Similar articles
-
Crystal structure of the protealysin precursor: insights into propeptide function.J Biol Chem. 2010 Jan 15;285(3):2003-13. doi: 10.1074/jbc.M109.015396. Epub 2009 Nov 13. J Biol Chem. 2010. PMID: 19915005 Free PMC article.
-
Crystal structure of an oligomer of proteolytic zymogens: detailed conformational analysis of the bovine ternary complex and implications for their activation.J Mol Biol. 1997 Jun 27;269(5):861-80. doi: 10.1006/jmbi.1997.1040. J Mol Biol. 1997. PMID: 9223647
-
Autoprocessing of prothiolsubtilisin E in which active-site serine 221 is altered to cysteine.J Biol Chem. 1994 Feb 11;269(6):4169-74. J Biol Chem. 1994. PMID: 8307978
-
Functional and structural insights into astacin metallopeptidases.Biol Chem. 2012 Oct;393(10):1027-41. doi: 10.1515/hsz-2012-0149. Biol Chem. 2012. PMID: 23092796 Review.
-
Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes.Protein Sci. 1998 Apr;7(4):815-36. doi: 10.1002/pro.5560070401. Protein Sci. 1998. PMID: 9568890 Free PMC article. Review.
Cited by
-
Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.Biochemistry. 2011 Oct 18;50(41):8862-79. doi: 10.1021/bi201118z. Epub 2011 Sep 26. Biochemistry. 2011. PMID: 21928835 Free PMC article.
-
Role of YpeB in cortex hydrolysis during germination of Bacillus anthracis spores.J Bacteriol. 2014 Oct;196(19):3399-409. doi: 10.1128/JB.01899-14. Epub 2014 Jul 14. J Bacteriol. 2014. PMID: 25022853 Free PMC article.
-
Identification of a chitinase-modifying protein from Fusarium verticillioides: truncation of a host resistance protein by a fungalysin metalloprotease.J Biol Chem. 2011 Oct 14;286(41):35358-35366. doi: 10.1074/jbc.M111.279646. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878653 Free PMC article.
-
Functional characterization of a subtilisin-like serine protease from Vibrio cholerae.J Biol Chem. 2019 Jun 21;294(25):9888-9900. doi: 10.1074/jbc.RA119.007745. Epub 2019 May 10. J Biol Chem. 2019. PMID: 31076508 Free PMC article.
-
A halophilic metalloprotease from Salinivibrio sp. YH4 and its application in antioxidant peptide production.Front Microbiol. 2025 May 19;16:1595109. doi: 10.3389/fmicb.2025.1595109. eCollection 2025. Front Microbiol. 2025. PMID: 40458704 Free PMC article.
References
-
- Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. 2nd Ed. Boston: Academic; 2004. pp. 374–399.
-
- Ohta Y, Ogura Y, Wada A. Thermostable protease from thermophilic bacteria. I. Thermostability, physiocochemical properties, and amino acid composition. J Biol Chem. 1966;241:5919–5925. - PubMed
-
- Titani K, Hermodson MA, Ericsson LH, Walsh KA, Neurath H. Amino-acid sequence of thermolysin. Nat New Biol. 1972;238:35–37. - PubMed
-
- Matthews BW, Jansonius JN, Colman PM, Schoenborn BP, Dupourque D. Three-dimensional structure of thermolysin. Nat New Biol. 1972;238:37–41. - PubMed
-
- Tronrud DE, Holden HM, Matthews BW. Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond. Science. 1987;235:571–574. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

