Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum
- PMID: 20876295
- PMCID: PMC2981305
- DOI: 10.1128/IAI.00834-10
Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum
Abstract
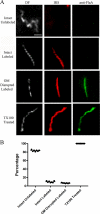
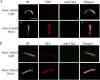
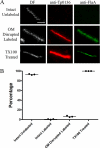
Treponema pallidum reacts poorly with the antibodies present in rabbit and human syphilitic sera, a property attributed to the paucity of proteins in its outer membrane. To better understand the basis for the syphilis spirochete's "stealth pathogenicity," we used a dual-label, 3-step amplified assay in which treponemes encapsulated in gel microdroplets were probed with syphilitic sera in parallel with anti-FlaA antibodies. A small (approximately 5 to 10%) but reproducible fraction of intact treponemes bound IgG and/or IgM antibodies. Three lines of evidence supported the notion that the surface antigens were likely β-barrel-forming outer membrane proteins (OMPs): (i) surface labeling with anti-lipoidal (VDRL) antibodies was not observed, (ii) immunoblot analysis confirmed prior results showing that T. pallidum glycolipids are not immunoreactive, and (iii) labeling of intact organisms was not appreciably affected by proteinase K (PK) treatment. With this method, we also demonstrate that TprK (TP0897), an extensively studied candidate OMP, and TP0136, a lipoprotein recently reported to be surface exposed, are both periplasmic. Consistent with the immunolabeling studies, TprK was also found to lack amphiphilicity, a characteristic property of β-barrel-forming proteins. Using a consensus computational framework that combined subcellular localization and β-barrel structural prediction tools, we generated ranked groups of candidate rare OMPs, the predicted T. pallidum outer membrane proteome (OMPeome), which we postulate includes the surface-exposed molecules detected by our enhanced gel microdroplet assay. In addition to underscoring the syphilis spirochete's remarkably poor surface antigenicity, our findings help to explain the complex and shifting balance between pathogen and host defenses that characterizes syphilitic infection.
Figures





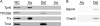

Similar articles
-
TprC/D (Tp0117/131), a trimeric, pore-forming rare outer membrane protein of Treponema pallidum, has a bipartite domain structure.J Bacteriol. 2012 May;194(9):2321-33. doi: 10.1128/JB.00101-12. Epub 2012 Mar 2. J Bacteriol. 2012. PMID: 22389487 Free PMC article.
-
Treponema pallidum in Gel Microdroplets: A Method for Topological Analysis of BamA (TP0326) and Localization of Rare Outer Membrane Proteins.Methods Mol Biol. 2015;1329:67-75. doi: 10.1007/978-1-4939-2871-2_6. Methods Mol Biol. 2015. PMID: 26427677
-
Structural Modeling of the Treponema pallidum Outer Membrane Protein Repertoire: a Road Map for Deconvolution of Syphilis Pathogenesis and Development of a Syphilis Vaccine.J Bacteriol. 2021 Jul 8;203(15):e0008221. doi: 10.1128/JB.00082-21. Epub 2021 Jul 8. J Bacteriol. 2021. PMID: 33972353 Free PMC article.
-
The Treponema pallidum Outer Membrane.Curr Top Microbiol Immunol. 2018;415:1-38. doi: 10.1007/82_2017_44. Curr Top Microbiol Immunol. 2018. PMID: 28849315 Free PMC article. Review.
-
Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema Pallidum Polypeptide Research Group.Microbiol Rev. 1993 Sep;57(3):750-79. doi: 10.1128/mr.57.3.750-779.1993. Microbiol Rev. 1993. PMID: 8246847 Free PMC article. Review.
Cited by
-
Investigation of the immune escape mechanism of Treponema pallidum.Infection. 2023 Apr;51(2):305-321. doi: 10.1007/s15010-022-01939-z. Epub 2022 Oct 19. Infection. 2023. PMID: 36260281 Review.
-
Two Potential Syphilis Vaccine Candidates Inhibit Dissemination of Treponema pallidum.Front Immunol. 2021 Nov 25;12:759474. doi: 10.3389/fimmu.2021.759474. eCollection 2021. Front Immunol. 2021. PMID: 34899710 Free PMC article.
-
Sequence Variation of Rare Outer Membrane Protein β-Barrel Domains in Clinical Strains Provides Insights into the Evolution of Treponema pallidum subsp. pallidum, the Syphilis Spirochete.mBio. 2018 Jun 12;9(3):e01006-18. doi: 10.1128/mBio.01006-18. mBio. 2018. PMID: 29895642 Free PMC article.
-
Syphilis.Nat Rev Dis Primers. 2017 Oct 12;3:17073. doi: 10.1038/nrdp.2017.73. Nat Rev Dis Primers. 2017. PMID: 29022569 Free PMC article. Review.
-
Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes.mBio. 2013 Feb 12;4(1):e00615-12. doi: 10.1128/mBio.00615-12. mBio. 2013. PMID: 23404400 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

