Nontransgenic genome modification in plant cells
- PMID: 20876340
- PMCID: PMC2971589
- DOI: 10.1104/pp.110.164806
Nontransgenic genome modification in plant cells
Abstract
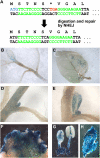
Zinc finger nucleases (ZFNs) are a powerful tool for genome editing in eukaryotic cells. ZFNs have been used for targeted mutagenesis in model and crop species. In animal and human cells, transient ZFN expression is often achieved by direct gene transfer into the target cells. Stable transformation, however, is the preferred method for gene expression in plant species, and ZFN-expressing transgenic plants have been used for recovery of mutants that are likely to be classified as transgenic due to the use of direct gene-transfer methods into the target cells. Here we present an alternative, nontransgenic approach for ZFN delivery and production of mutant plants using a novel Tobacco rattle virus (TRV)-based expression system for indirect transient delivery of ZFNs into a variety of tissues and cells of intact plants. TRV systemically infected its hosts and virus ZFN-mediated targeted mutagenesis could be clearly observed in newly developed infected tissues as measured by activation of a mutated reporter transgene in tobacco (Nicotiana tabacum) and petunia (Petunia hybrida) plants. The ability of TRV to move to developing buds and regenerating tissues enabled recovery of mutated tobacco and petunia plants. Sequence analysis and transmission of the mutations to the next generation confirmed the stability of the ZFN-induced genetic changes. Because TRV is an RNA virus that can infect a wide range of plant species, it provides a viable alternative to the production of ZFN-mediated mutants while avoiding the use of direct plant-transformation methods.
Figures






Similar articles
-
Targeted mutagenesis using zinc-finger nucleases in perennial fruit trees.Planta. 2015 Apr;241(4):941-51. doi: 10.1007/s00425-014-2224-x. Epub 2014 Dec 21. Planta. 2015. PMID: 25528147
-
Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis.Virus Res. 2018 Jan 15;244:333-337. doi: 10.1016/j.virusres.2017.10.009. Epub 2017 Oct 16. Virus Res. 2018. PMID: 29051052
-
Targeting DNA to a previously integrated transgenic locus using zinc finger nucleases.Methods Mol Biol. 2012;847:391-7. doi: 10.1007/978-1-61779-558-9_31. Methods Mol Biol. 2012. PMID: 22351024
-
Gene targeting in plants: 25 years later.Int J Dev Biol. 2013;57(6-8):629-37. doi: 10.1387/ijdb.130194hp. Int J Dev Biol. 2013. PMID: 24166445 Review.
-
Genome editing in plant cells by zinc finger nucleases.Trends Plant Sci. 2010 Jun;15(6):308-21. doi: 10.1016/j.tplants.2010.03.001. Epub 2010 Mar 26. Trends Plant Sci. 2010. PMID: 20347379 Review.
Cited by
-
TALENs: a widely applicable technology for targeted genome editing.Nat Rev Mol Cell Biol. 2013 Jan;14(1):49-55. doi: 10.1038/nrm3486. Epub 2012 Nov 21. Nat Rev Mol Cell Biol. 2013. PMID: 23169466 Free PMC article. Review.
-
Virus-induced genome editing using a miniature CRISPR system.Nat Plants. 2025 May;11(5):951-952. doi: 10.1038/s41477-025-01990-2. Nat Plants. 2025. PMID: 40269177 No abstract available.
-
A rapid assay to quantify the cleavage efficiency of custom-designed nucleases in planta.Plant Mol Biol. 2013 Jun;82(3):207-21. doi: 10.1007/s11103-013-0052-1. Epub 2013 Apr 28. Plant Mol Biol. 2013. PMID: 23625357
-
Genome Editing: A Promising Approach for Achieving Abiotic Stress Tolerance in Plants.Int J Genomics. 2022 Apr 15;2022:5547231. doi: 10.1155/2022/5547231. eCollection 2022. Int J Genomics. 2022. PMID: 35465040 Free PMC article. Review.
-
Plant Genome Engineering for Targeted Improvement of Crop Traits.Front Plant Sci. 2019 Feb 12;10:114. doi: 10.3389/fpls.2019.00114. eCollection 2019. Front Plant Sci. 2019. PMID: 30809237 Free PMC article. Review.
References
-
- Baribault H, Kemler R. (1989) Embryonic stem cell culture and gene targeting in transgenic mice. Mol Biol Med 6: 481–492 - PubMed
-
- Bernatzky R, Tanksley SD. (1986) Genetics of actin-related sequences in tomato. Theor Appl Genet 72: 314–321 - PubMed
-
- Britt AB, May GD. (2003) Re-engineering plant gene targeting. Trends Plant Sci 8: 90–95 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

