In situ detection of active transglutaminases for keratinocyte type (TGase 1) and tissue type (TGase 2) using fluorescence-labeled highly reactive substrate peptides
- PMID: 20876521
- PMCID: PMC3201132
- DOI: 10.1369/jhc.2010.957225
In situ detection of active transglutaminases for keratinocyte type (TGase 1) and tissue type (TGase 2) using fluorescence-labeled highly reactive substrate peptides
Abstract
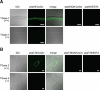
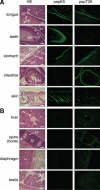
Transglutaminase is a calcium-dependent enzyme that posttranslationally modifies proteins by cross-linking between glutamine and lysine residues or attachment of a primary amine to specific polypeptide-bound glutamine residues. Eight isozymes play essential roles in various mammalian biological processes. The authors have recently identified 12–amino acid preferred substrate peptide sequences that are highly reactive and act in an isozyme-specific manner. In this study, a rapid, isozyme-specific, and sensitive detection of active keratinocyte type (TGase 1) and tissue type (TGase 2) was successful using fluorescence-labeled peptides. This procedure involved using whole-body sections of a mouse to extensively analyze the tissue distribution of both enzymes that revealed clearly distinct patterns. Strong active TGase 1 was observed in epithelial tissues such as tongue, developing teeth, forestomach, and skin epidermis. Significantly active TGase 2 was observed in various types of tissues as predicted and at particularly higher levels in the intestinal mucosa, muscle membrane, and whole veins in the liver. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures


Similar articles
-
Identification of a preferred substrate peptide for transglutaminase 3 and detection of in situ activity in skin and hair follicles.FEBS J. 2010 Sep;277(17):3564-74. doi: 10.1111/j.1742-4658.2010.07765.x. FEBS J. 2010. PMID: 20716179
-
Identification of preferred substrate sequences for transglutaminase 1--development of a novel peptide that can efficiently detect cross-linking enzyme activity in the skin.FEBS J. 2008 Nov;275(22):5667-77. doi: 10.1111/j.1742-4658.2008.06692.x. FEBS J. 2008. PMID: 18959752
-
Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: identification of peptide substrates for TGASE 2 and Factor XIIIA.J Biol Chem. 2006 Jun 30;281(26):17699-706. doi: 10.1074/jbc.M513538200. Epub 2006 Apr 24. J Biol Chem. 2006. PMID: 16636049
-
Identification and characterization of substrates crosslinked by transglutaminases in liver and kidney fibrosis.Anal Biochem. 2020 Sep 1;604:113629. doi: 10.1016/j.ab.2020.113629. Epub 2020 Feb 13. Anal Biochem. 2020. PMID: 32061735 Review.
-
Preferred substrate sequences for transglutaminase 2: screening using a phage-displayed peptide library.Amino Acids. 2009 Apr;36(4):619-24. doi: 10.1007/s00726-008-0126-6. Epub 2008 Jul 24. Amino Acids. 2009. PMID: 18651094 Review.
Cited by
-
Niche-specific requirement for hyphal wall protein 1 in virulence of Candida albicans.PLoS One. 2013 Nov 8;8(11):e80842. doi: 10.1371/journal.pone.0080842. eCollection 2013. PLoS One. 2013. PMID: 24260489 Free PMC article.
-
Global identification and analysis of isozyme-specific possible substrates crosslinked by transglutaminases using substrate peptides in mouse liver fibrosis.Sci Rep. 2017 Mar 22;7:45049. doi: 10.1038/srep45049. Sci Rep. 2017. PMID: 28327670 Free PMC article.
-
Biocatalysis by Transglutaminases: A Review of Biotechnological Applications.Micromachines (Basel). 2018 Oct 31;9(11):562. doi: 10.3390/mi9110562. Micromachines (Basel). 2018. PMID: 30715061 Free PMC article. Review.
-
Biochemical Characterization of Medaka (Oryzias latipes) Transglutaminases, OlTGK1 and OlTGK2, as Orthologues of Human Keratinocyte-Type Transglutaminase.PLoS One. 2015 Dec 29;10(12):e0144194. doi: 10.1371/journal.pone.0144194. eCollection 2015. PLoS One. 2015. PMID: 26713442 Free PMC article.
-
Isozyme-specific comprehensive characterization of transglutaminase-crosslinked substrates in kidney fibrosis.Sci Rep. 2018 May 9;8(1):7306. doi: 10.1038/s41598-018-25674-4. Sci Rep. 2018. PMID: 29743665 Free PMC article.
References
-
- Aeschlimann D, Koeller MK, Allen-Hoffmann BL, Mosher DF. 1998. Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes: detection and identification of transglutaminase gene products based on reverse transcription-polymerase chain reaction with degenerate primers. J Biol Chem. 273:3452-3460 - PubMed
-
- Beninati S, Piacentini M. 2004. The transglutaminase family: an overview. Amino Acids. 26:367-372 - PubMed
-
- Candi E, Schmidt R, Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 6:328-340 - PubMed
-
- Eckert RL, Sturniolo MT, Broome AM, Ruse M, Rorke EA. 2005. Transglutaminase function in epidermis. J Invest Dermatol. 124:481-492 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

