Structural characterization of naturally occurring RNA single mismatches
- PMID: 20876693
- PMCID: PMC3035445
- DOI: 10.1093/nar/gkq793
Structural characterization of naturally occurring RNA single mismatches
Abstract
RNA is known to be involved in several cellular processes; however, it is only active when it is folded into its correct 3D conformation. The folding, bending and twisting of an RNA molecule is dependent upon the multitude of canonical and non-canonical secondary structure motifs. These motifs contribute to the structural complexity of RNA but also serve important integral biological functions, such as serving as recognition and binding sites for other biomolecules or small ligands. One of the most prevalent types of RNA secondary structure motifs are single mismatches, which occur when two canonical pairs are separated by a single non-canonical pair. To determine sequence-structure relationships and to identify structural patterns, we have systematically located, annotated and compared all available occurrences of the 30 most frequently occurring single mismatch-nearest neighbor sequence combinations found in experimentally determined 3D structures of RNA-containing molecules deposited into the Protein Data Bank. Hydrogen bonding, stacking and interaction of nucleotide edges for the mismatched and nearest neighbor base pairs are described and compared, allowing for the identification of several structural patterns. Such a database and comparison will allow researchers to gain insight into the structural features of unstudied sequences and to quickly look-up studied sequences.
Figures

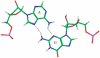
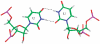
 in the hydrogen bonded, stacked orientation (PDB ID 1O9M) (a) and in the non-hydrogen bonded, unstacked orientation (PDB ID 1O9M) (b).
in the hydrogen bonded, stacked orientation (PDB ID 1O9M) (a) and in the non-hydrogen bonded, unstacked orientation (PDB ID 1O9M) (b).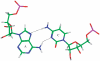
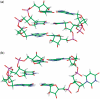
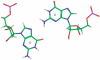
 . This mismatch-nearest neighbor sequence combination is found in the 30 most frequently occurring single mismatches (84) and accounts for 80% of the total A·C mismatches found in this study.
. This mismatch-nearest neighbor sequence combination is found in the 30 most frequently occurring single mismatches (84) and accounts for 80% of the total A·C mismatches found in this study.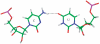
 (84) that was also represented in the PDB.
(84) that was also represented in the PDB.Similar articles
-
Thermodynamic characterization of single mismatches found in naturally occurring RNA.Biochemistry. 2007 Nov 20;46(46):13425-36. doi: 10.1021/bi701311c. Epub 2007 Oct 24. Biochemistry. 2007. PMID: 17958380
-
Thermodynamics of unpaired terminal nucleotides on short RNA helixes correlates with stacking at helix termini in larger RNAs.J Mol Biol. 1999 Jul 30;290(5):967-82. doi: 10.1006/jmbi.1999.2906. J Mol Biol. 1999. PMID: 10438596
-
Thermodynamics of single mismatches in RNA duplexes.Biochemistry. 1999 Oct 26;38(43):14214-23. doi: 10.1021/bi991186l. Biochemistry. 1999. PMID: 10571995
-
The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems.EMBO Rep. 2000 Jul;1(1):18-23. doi: 10.1093/embo-reports/kvd001. EMBO Rep. 2000. PMID: 11256617 Free PMC article. Review.
-
RNA structure and dynamics: a base pairing perspective.Prog Biophys Mol Biol. 2013 Nov;113(2):264-83. doi: 10.1016/j.pbiomolbio.2013.07.003. Epub 2013 Jul 23. Prog Biophys Mol Biol. 2013. PMID: 23891726 Review.
Cited by
-
pH dependence of C•A, G•A and A•A mismatches in the stem of precursor microRNA-31.Biophys Chem. 2022 Apr;283:106763. doi: 10.1016/j.bpc.2022.106763. Epub 2022 Jan 22. Biophys Chem. 2022. PMID: 35114594 Free PMC article.
-
RNA tertiary structure and conformational dynamics revealed by BASH MaP.bioRxiv [Preprint]. 2024 Aug 19:2024.04.11.589009. doi: 10.1101/2024.04.11.589009. bioRxiv. 2024. Update in: Elife. 2024 Dec 03;13:RP98540. doi: 10.7554/eLife.98540. PMID: 38645201 Free PMC article. Updated. Preprint.
-
Ensemble analysis of primary microRNA structure reveals an extensive capacity to deform near the Drosha cleavage site.Biochemistry. 2013 Feb 5;52(5):795-807. doi: 10.1021/bi301452a. Epub 2013 Jan 18. Biochemistry. 2013. PMID: 23305493 Free PMC article.
-
Electronic Circular Dichroism of the Cas9 Protein and gRNA:Cas9 Ribonucleoprotein Complex.Int J Mol Sci. 2021 Mar 13;22(6):2937. doi: 10.3390/ijms22062937. Int J Mol Sci. 2021. PMID: 33805827 Free PMC article.
-
Effect of sodium ions on RNA duplex stability.Biochemistry. 2013 Oct 22;52(42):7477-85. doi: 10.1021/bi4008275. Epub 2013 Oct 9. Biochemistry. 2013. PMID: 24106785 Free PMC article.
References
-
- Mao H, White SA, Williamson JR. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 1999;6:1139–1147. - PubMed
-
- Beuth B, García-Mayoral MF, Taylor IA, Ramos A. Scaffold-independent analysis of RNA-protein interactions: the nova-1 KH3-RNA complex. J. Am. Chem. Soc. 2007;129:10205–10210. - PubMed
-
- Messias AC, Sattler M. Structural basis of single-stranded RNA recognition. Acc. Chem. Res. 2004;37:279–287. - PubMed

