A novel mechanism of acid and bile acid-induced DNA damage involving Na+/H+ exchanger: implication for Barrett's oesophagus
- PMID: 20876775
- PMCID: PMC3087378
- DOI: 10.1136/gut.2010.213686
A novel mechanism of acid and bile acid-induced DNA damage involving Na+/H+ exchanger: implication for Barrett's oesophagus
Abstract
Objective: Barrett's oesophagus is a premalignant disease associated with oesophageal adenocarcinoma. The major goal of this study was to determine the mechanism responsible for bile acid-induced alteration in intracellular pH (pH(i)) and its effect on DNA damage in cells derived from normal oesophagus (HET1A) or Barrett's oesophagus (CP-A).
Design: Cells were exposed to bile acid cocktail (BA) and/or acid in the presence/absence of inhibitors of nitric oxide synthase (NOS) or sodium-hydrogen exchanger (NHE). Nitric oxide (NO), pH(i) and DNA damage were measured by fluorescent imaging and comet assay. Expression of NHE1 and NOS in cultured cells and biopsies from Barrett's oesophagus or normal squamous epithelium was determined by RT-PCR, immunoblotting or immunohistochemistry.
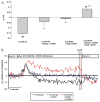
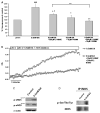
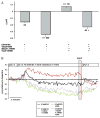
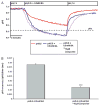
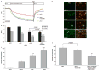
Results: A dose dependent decrease in pH(i) was observed in CP-A cells exposed to BA. This effect of BA is the consequence of NOS activation and increased NO production, which leads to NHE inhibition. Exposure of oesophageal cells to acid in combination with BA synergistically decreased pH(i). The decrease was more pronounced in CP-A cells and resulted in >2-fold increase in DNA damage compared to acid treatment alone. Examination of biopsies and cell lines revealed robust expression of NHE1 in Barrett's oesophagus but an absence of NHE1 in normal epithelium.
Conclusions: The results of this study identify a new mechanism of bile acid-induced DNA damage. We found that bile acids present in the refluxate activate immediately all three isoforms of NOS, which leads to an increased NO production and NHE inhibition. This consequently results in increased intracellular acidification and DNA damage, which may lead to mutations and cancer progression. Therefore, we propose that in addition to gastric reflux, bile reflux should be controlled in patients with Barrett's oesophagus.
Conflict of interest statement
Figures



References
-
- Rastogi A, Puli S, El-Serag HB, et al. Incidence of esophageal adenocarcinoma in patients with Barrett’s oesophagus and high-grade dysplasia: a meta-analysis. Gastrointest Endosc. 2008;67:394–8. - PubMed
-
- Menges M, Muller M, Zeitz M. Increased acid and bile reflux in Barrett’s oesophagus compared to reflux esophagitis, and effect of proton pump inhibitor therapy. Am J Gastroenterol. 2001;96:331–7. - PubMed
-
- Ostrowski J, Mikula M, Karczmarski J, et al. Molecular defense mechanisms of Barrett’s metaplasia estimated by an integrative genomics. J Mol Med. 2007;85:733–43. - PubMed
-
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–18. - PubMed
-
- LeBoeuf RA, Kerckaert GA. The induction of transformed-like morphology and enhanced growth in Syrian hamster embryo cells grown at acidic pH. Carcinogenesis. 1986;7:1431–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
