Activation-induced cytidine deaminase accelerates clonal evolution in BCR-ABL1-driven B-cell lineage acute lymphoblastic leukemia
- PMID: 20876806
- PMCID: PMC2948648
- DOI: 10.1158/0008-5472.CAN-10-1438
Activation-induced cytidine deaminase accelerates clonal evolution in BCR-ABL1-driven B-cell lineage acute lymphoblastic leukemia
Abstract
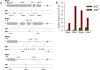
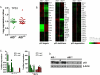
Activation-induced cytidine deaminase (AID) is required for somatic hypermutation and immunoglobulin (Ig) class switch recombination in germinal center (GC) B cells. Occasionally, AID can target non-Ig genes and thereby promote GC B-cell lymphomagenesis. We recently showed that the oncogenic BCR-ABL1 kinase induces aberrant expression of AID in pre-B acute lymphoblastic leukemia (ALL) and lymphoid chronic myelogenous leukemia blast crisis. To elucidate the biological significance of aberrant AID expression, we studied loss of AID function in a murine model of BCR-ABL1 ALL. Mice transplanted with BCR-ABL1-transduced AID(-/-) bone marrow had prolonged survival compared with mice transplanted with leukemia cells generated from AID(+/+) bone marrow. Consistent with a causative role of AID in genetic instability, AID(-/-) leukemia had a lower frequency of amplifications and deletions and a lower frequency of mutations in non-Ig genes, including Pax5 and Rhoh compared with AID(+/+) leukemias. AID(-/-) and AID(+/+) ALL cells showed a markedly distinct gene expression pattern, and AID(-/-) ALL cells failed to downregulate a number of tumor-suppressor genes including Rhoh, Cdkn1a (p21), and Blnk (SLP65). We conclude that AID accelerates clonal evolution in BCR-ABL1 ALL by enhancing genetic instability and aberrant somatic hypermutation, and by negative regulation of tumor-suppressor genes.
© 2010 AACR.
Figures
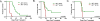


References
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. - PubMed
-
- Ramiro A, San-Martin BR, McBride K, et al. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv. Immunol. 2007;94:75–107. - PubMed
-
- Liu M, Duke JL, Richter DJ, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. - PubMed
-
- Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

