A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes
- PMID: 20876833
- PMCID: PMC2957065
- DOI: 10.1261/rna.2091710
A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes
Abstract
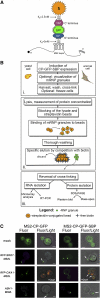
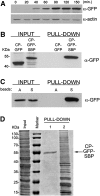
Intracellular mRNA targeting and localized translation are potential determinants for protein localization. To facilitate targeting, mRNAs possess specific cis-acting sequence motifs that are recognized by trans-acting RNA-binding proteins (RBPs). While many mRNAs are trafficked, our knowledge of the RBPs involved and presence of additional transcripts within these ribonucleoprotein (RNP) complexes is limited. To facilitate the identification of RBPs and transcripts that bind to specific mRNAs, we developed RNA-binding protein purification and identification (RaPID), a novel technique that allows for the affinity purification of MS2 aptamer-tagged mRNAs and subsequent detection of bound RBPs and transcripts using mass-spectometry and RT-PCR, respectively. RaPID effectively isolated specific mRNAs from both yeast and mammalian cells, and identified known mRNA-RBP interactions (e.g., ASH1-She2; β-Actin-IMP1). By isolating tagged OXA1 mRNA using RaPID, we could identify a yeast COPI subunit (i.e., Sec27) as a candidate interacting protein. This finding was strengthened by the observation that a portion of OXA1 mRNA was delocalized in a sec27-1 temperature-sensitive mutant at restrictive temperatures. Finally, RaPID could also be used to show biochemically the coexistence of different RNA species within the same RNP complex (e.g., coprecipitation of the yeast SRO7, WSC2, SEC3, and IST2 mRNAs with ASH1 mRNA) for the first time.
Figures
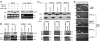
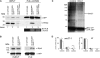


References
-
- Aronov S, Gerst JE 2004. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J Biol Chem 279: 36962–36971 - PubMed
-
- Beach DL, Keene JD 2008. Ribotrap : Targeted purification of RNA-specific RNPs from cell lysates through immunoaffinity precipitation to identify regulatory proteins and RNAs. Methods Mol Biol 419: 69–91 - PubMed
-
- Beach DL, Salmon ED, Bloom K 1999. Localization and anchoring of mRNA in budding yeast. Curr Biol 9: 569–578 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
