Cobalamin- and corrinoid-dependent enzymes
- PMID: 20877792
- PMCID: PMC3120101
- DOI: 10.1039/BK9781847559159-00053
Cobalamin- and corrinoid-dependent enzymes
Abstract
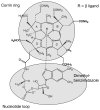
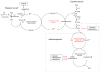
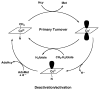
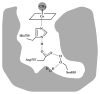
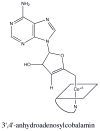
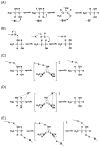
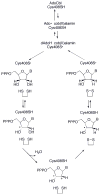
This chapter reviews the literature on cobalamin- and corrinoid-containing enzymes. These enzymes fall into two broad classes, those using methylcobalamin or related methylcorrinoids as prosthetic groups and catalyzing methyl transfer reactions, and those using adenosylcobalamin as the prosthetic group and catalyzing the generation of substrate radicals that in turn undergo rearrangements and/or eliminations.
Figures
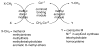
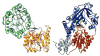
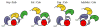





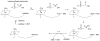



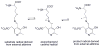
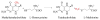

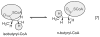
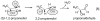
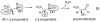
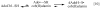



References
-
- Ragsdale SW, Lindahl PA, Munck E. J Biol Chem. 1987;262:14289–14297. - PubMed
-
- Stupperich E, Eisenger HJ, Albracht SPJ. Eur J Biochem. 1990;193:105–109. - PubMed
-
- Ragsdale SW. In: Chemistry and biochemistry of B12. Banerjee R, editor. John Wiley; New York: 1999. pp. 633–653.
-
- Banerjee RV, Johnston NL, Sobeski JK, Datta P, Matthews RG. J Biol Chem. 1989;264:13888–13895. - PubMed
-
- Amaratunga M, Fluhr K, Jarrett JT, Drennan CL, Ludwig ML, Matthews RG, Scholten JD. Biochemistry. 1996;35:2453–2463. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
