APOBEC2, a selective inhibitor of TGFβ signaling, regulates left-right axis specification during early embryogenesis
- PMID: 20880495
- PMCID: PMC3038383
- DOI: 10.1016/j.ydbio.2010.09.016
APOBEC2, a selective inhibitor of TGFβ signaling, regulates left-right axis specification during early embryogenesis
Abstract
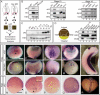
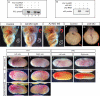
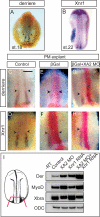
The specification of left-right asymmetry is an evolutionarily conserved developmental process in vertebrates. The interplay between two TGFβ ligands, Derrière/GDF1 and Xnr1/Nodal, together with inhibitors such as Lefty and Coco/Cerl2, have been shown to provide the signals that lead to the establishment of laterality. However, molecular events leading to and following these signals remain mostly unknown. We find that APOBEC2, a member of the cytidine deaminase family of DNA/RNA editing enzymes, is induced by TGFβ signaling, and that its activity is necessary to specify the left-right axis in Xenopus and zebrafish embryos. Surprisingly, we find that APOBEC2 selectively inhibits Derrière, but not Xnr1, signaling. The inhibitory effect is conserved, as APOBEC2 blocks TGFβ signaling, and promotes muscle differentiation, in a mammalian myoblastic cell line. This demonstrates for the first time that a putative RNA/DNA editing enzyme regulates TGFβ signaling and plays a major role in development.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Anant S, Mukhopadhyay D, Sankaranand V, Kennedy S, Henderson JO, Davidson NO. ARCD-1, an apobec-1-related cytidine deaminase, exerts a dominant negative effect on C to U RNA editing. Am J Physiol Cell Physiol. 2001;281:C1904–16. - PubMed
-
- Artaza JN, Bhasin S, Mallidis C, Taylor W, Ma K, Gonzalez-Cadavid NF. Endogenous expression and localization of myostatin and its relation to myosin heavy chain distribution in C2C12 skeletal muscle cells. J Cell Physiol. 2002;190:170–9. - PubMed
-
- Bell E, Munoz-Sanjuan I, Altmann CR, Vonica A, Brivanlou AH. Cell fate specification and competence by Coco, a maternal BMP, TGFbeta and Wnt inhibitor. Development. 2003;130:1381–9. - PubMed
-
- Bisgrove BW, Yost HJ. Classification of left-right patterning defects in zebrafish, mice, and humans. American Journal of Medical Genetics. 2001;101:315–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

