Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin
- PMID: 20880836
- PMCID: PMC2955093
- DOI: 10.1073/pnas.1004361107
Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin
Abstract
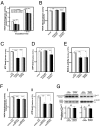
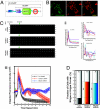
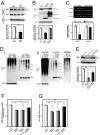
Disrupted-in-schizophrenia 1 (DISC1) has emerged as a schizophrenia-susceptibility gene affecting various neuronal functions. In this study, we characterized Mitofilin, a mitochondrial inner membrane protein, as a mediator of the mitochondrial function of DISC1. A fraction of DISC1 was localized to the inside of mitochondria and directly interacts with Mitofilin. A reduction in DISC1 function induced mitochondrial dysfunction, evidenced by decreased mitochondrial NADH dehydrogenase activities, reduced cellular ATP contents, and perturbed mitochondrial Ca(2+) dynamics. In addition, deficiencies in DISC1 and Mitofilin induced a reduction in mitochondrial monoamine oxidase-A activity. The mitochondrial dysfunctions evoked by the deficiency of DISC1 were partially phenocopied by an overexpression of truncated DISC1 that is associated with schizophrenia in human. DISC1 deficiencies induced the ubiquitination of Mitofilin, suggesting that DISC1 is critical for the stability of Mitofilin. Finally, the mitochondrial dysfunction induced by DISC1 deficiency was partially reversed by coexpression of Mitofilin, confirming a functional link between DISC1 and Mitofilin for the normal mitochondrial function. According to these results, we propose that DISC1 plays essential roles for mitochondrial function in collaboration with a mitochondrial interacting partner, Mitofilin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- American Psychiatric Association—Task Force on DSM-IV . Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. 4th Ed. Washington, DC: American Psychiatric Association; 2000. p. xxxvii.
-
- Cardno AG, Gottesman II. Twin studies of schizophrenia: From bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. - PubMed
-
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry. 2005;10:40–68. - PubMed
-
- Mackie S, Millar JK, Porteous DJ. Role of DISC1 in neural development and schizophrenia. Curr Opin Neurobiol. 2007;17:95–102. - PubMed
-
- Kamiya A, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

