Comparison of E1A CR3-dependent transcriptional activation across six different human adenovirus subgroups
- PMID: 20881041
- PMCID: PMC3004344
- DOI: 10.1128/JVI.01243-10
Comparison of E1A CR3-dependent transcriptional activation across six different human adenovirus subgroups
Abstract
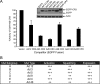
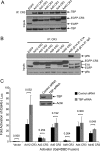
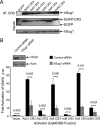
The largest E1A isoform of human adenovirus (Ad) includes a C-4 zinc finger domain within conserved region 3 (CR3) that is largely responsible for activating transcription of the early viral genes. CR3 interacts with multiple cellular factors, but its mechanism of action is modeled primarily on the basis of the mechanism for the prototype E1A protein of human Ad type 5. We expanded this model to include a representative member from each of the six human Ad subgroups. All CR3 domains tested were capable of transactivation. However, there were dramatic differences in their levels of transcriptional activation. Despite these functional variations, the interactions of these representative CR3s with known cellular transcriptional regulators revealed only modest differences. Four common cellular targets of all representative CR3s were identified: the proteasome component human Sug1 (hSug1)/S8, the acetyltransferases p300/CREB binding protein (CBP), the mediator component mediator complex subunit 23 (MED23) protein, and TATA binding protein (TBP). The first three factors appear to be critical for CR3 function. RNA interference against human TBP showed no significant reduction in transactivation by any CR3 tested. These results indicate that the cellular factors previously shown to be important for transactivation by Ad5 CR3 are similarly bound by the E1A proteins of other types. This was confirmed experimentally using a transcriptional squelching assay, which demonstrated that the CR3 regions of each Ad type could compete with Ad5 CR3 for limiting factors. Interestingly, a mutant of Ad5 CR3 (V147L) was capable of squelching wild-type Ad5 CR3, despite its failure to bind TBP, MED23, p300/CBP-associated factor (pCAF), or p300/CBP, suggestive of the possibility that an additional as yet unidentified cellular factor is required for transactivation by E1A CR3.
Figures





References
-
- Avvakumov, N., A. E. Kajon, R. C. Hoeben, and J. S. Mymryk. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329:477-492. - PubMed
-
- Avvakumov, N., J. Torchia, and J. S. Mymryk. 2003. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 22:3833-3841. - PubMed
-
- Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673-7685. - PubMed
-
- Berk, A. J., F. Lee, T. Harrison, J. Williams, and P. A. Sharp. 1979. A pre-early adenovirus 5 gene product regulates synthesis of early messenger RNAs. Cell 17:935-944. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

