Nuclear organization and dynamics of 7SK RNA in regulating gene expression
- PMID: 20881057
- PMCID: PMC2993747
- DOI: 10.1091/mbc.E10-02-0105
Nuclear organization and dynamics of 7SK RNA in regulating gene expression
Abstract
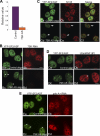
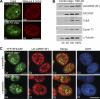
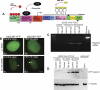
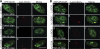
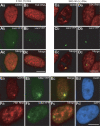
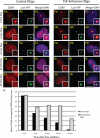
Noncoding RNAs play important roles in various aspects of gene regulation. We have identified 7SK RNA to be enriched in nuclear speckles or interchromatin granule clusters (IGCs), a subnuclear domain enriched in pre-mRNA processing factors. 7SK RNA, in association with HEXIM 1 and 2, is involved in the inhibition of transcriptional elongation by RNA polymerase II. Inhibition occurs via sequestration of the active P-TEFb kinase complex (CDK 9 and Cyclin T1/T2a/b or K) that is involved in phosphorylating the C-terminal domain of RNA polymerase II. Our results demonstrate that knock-down of 7SK RNA, by specific antisense oligonucleotides, results in the mislocalization of nuclear speckle constituents in a transcription-dependent manner, and the transcriptional up-regulation of a RNA polymerase II transcribed reporter gene locus. Furthermore, 7SK RNA transiently associates with a stably integrated reporter gene locus upon transcriptional down-regulation and its presence correlates with the efficient displacement of P-TEFb constituents from the locus. Our results suggest that 7SK RNA plays a role in modulating the available level of P-TEFb upon transcriptional down-regulation by sequestering its constituents in nuclear speckles.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

