Lipoic acid synthesis: a new family of octanoyltransferases generally annotated as lipoate protein ligases
- PMID: 20882995
- PMCID: PMC2982868
- DOI: 10.1021/bi101215f
Lipoic acid synthesis: a new family of octanoyltransferases generally annotated as lipoate protein ligases
Abstract
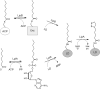
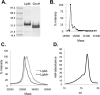
Bacillus subtilis lacks a recognizable homologue of the LipB octanoyltransferase, an enzyme essential for lipoic acid synthesis in Escherichia coli. LipB transfers the octanoyl moiety from octanoyl-acyl carrier protein to the lipoyl domains of the 2-oxoacid dehydrogenases via a thioester-linked octanoyl-LipB intermediate. The octanoylated dehydrogenase is then converted to the enzymatically active lipoylated species by insertion of two sulfur atoms into the octanoyl moiety by the S-adenosyl-l-methionine radical enzyme, LipA (lipoate synthase). B. subtilis synthesizes lipoic acid and contains a LipA homologue that is fully functional in E. coli. Therefore, the lack of a LipB homologue presented the puzzle of how B. subtilis synthesizes the LipA substrate. We report that B. subtilis encodes an octanoyltransferase that has virtually no sequence resemblance to E. coli LipB but instead has a sequence that resembles that of the E. coli lipoate ligase, LplA. On the basis of this resemblance, these genes have generally been annotated as encoding a lipoate ligase, an enzyme that in E. coli scavenges lipoic acid from the environment but plays no role in de novo synthesis. We have named the B. subtilis octanoyltransferase LipM and find that, like LipB, the LipM reaction proceeds through a thioester-linked acyl enzyme intermediate. The LipM active site nucleophile was identified as C150 by the finding that this thiol becomes modified when LipM is expressed in E. coli. The level of the octanoyl-LipM intermediate can be significantly decreased by blocking fatty acid synthesis during LipM expression, and C150 was confirmed as an essential active site residue by site-directed mutagenesis. LipM homologues seem the sole type of octanoyltransferase present in the firmicutes and are also present in the cyanobacteria. LipM type octanoyltransferases represent a new clade of the PF03099 protein family, suggesting that octanoyl transfer activity has evolved at least twice within this superfamily.
Figures
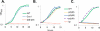

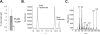

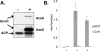

References
-
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 2000;69:961–1004. - PubMed
-
- Xiao Z, Xu P. Acetoin metabolism in bacteria. Crit Rev Microbiol. 2007;33:127–140. - PubMed
-
- Cronan JE, Zhao X, Jiang Y. Function, attachment and synthesis of lipoic acid in Escherichia coli. Advances in Microbial Physiology. 2005:103–146. - PubMed
-
- Zhao X, Miller JR, Cronan JE. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry. 2005;44:16737–16746. - PubMed
-
- Douglas P, Kriek M, Bryant P, Roach PL. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew Chem Int Ed Engl. 2006;45:5197–5199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

