Oocyte spindle proteomics analysis leading to rescue of chromosome congression defects in cloned embryos
- PMID: 20883044
- PMCID: PMC4316211
- DOI: 10.1021/pr100827j
Oocyte spindle proteomics analysis leading to rescue of chromosome congression defects in cloned embryos
Abstract
Embryos produced by somatic cell nuclear transfer (SCNT) display low term developmental potential. This is associated with deficiencies in spindle composition prior to activation and at early mitotic divisions, including failure to assemble certain proteins on the spindle. The protein-deficient spindles are accompanied by chromosome congression defects prior to activation and during the first mitotic divisions of the embryo. The molecular basis for these deficiencies and how they might be avoided are unknown. Proteomic analyses of spindles isolated from normal metaphase II (MII) stage oocytes and SCNT constructs, along with a systematic immunofluorescent survey of known spindle-associated proteins were undertaken. This was the first proteomics study of mammalian oocyte spindles. The study revealed four proteins as being deficient in spindles of SCNT embryos in addition to those previously identified; these were clathrin heavy chain (CLTC), aurora B kinase, dynactin 4, and casein kinase 1 alpha. Due to substantial reduction in CLTC abundance after spindle removal, we undertook functional studies to explore the importance of CLTC in oocyte spindle function and in chromosome congression defects of cloned embryos. Using siRNA knockdown, we demonstrated an essential role for CLTC in chromosome congression during oocyte maturation. We also demonstrated rescue of chromosome congression defects in SCNT embryos at the first mitosis using CLTC mRNA injection. These studies are the first to employ proteomics analyses coupled to functional interventions to rescue a specific molecular defect in cloned embryos.
Figures
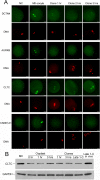
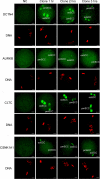
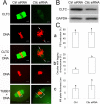
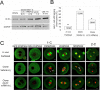
References
-
- Miyara F, Han Z, Gao S, Vassena R, Latham KE. Non-equivalence of embryonic and somatic cell nuclei affecting spindle composition in clones. Dev Biol. 2006;289(1):206–17. - PubMed
-
- Simerly C, Dominko T, Navara C, Payne C, Capuano S, Gosman G, Chong KY, Takahashi D, Chace C, Compton D, Hewitson L, Schatten G. Molecular correlates of primate nuclear transfer failures. Science. 2003;300(5617):297. - PubMed
-
- Chung YG, Ratnam S, Chaillet JR, Latham KE. Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol Reprod. 2003;69(1):146–53. - PubMed
-
- Gao S, Chung YG, Williams JW, Riley J, Moley K, Latham KE. Somatic cell-like features of cloned mouse embryos prepared with cultured myoblast nuclei. Biol Reprod. 2003;69(1):48–56. - PubMed
-
- Gao S, McGarry M, Latham KE, Wilmut I. Cloning of mice by nuclear transfer. Cloning Stem Cells. 2003;5(4):287–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

