Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities
- PMID: 20883224
- PMCID: PMC2951289
- DOI: 10.1111/j.2041-1014.2010.00585.x
Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities
Abstract
More than 700 bacterial species have been detected in the human oral cavity. They form highly organized microbial communities and are responsible for many oral infectious diseases, such as dental caries and periodontal disease. The prevention and treatment of these diseases require a comprehensive knowledge of oral microbial communities, which largely relies on culture-dependent methods to provide detailed phenotypic and physiological analysis of these communities. However, most of the currently available laboratory media can only selectively support the growth of a limited number of bacterial species within these communities, and fail to sustain the original oral microbial diversity. In this study, using denaturing gradient gel electrophoresis (DGGE) as an index to systematically survey and analyse the selectivity of commonly used laboratory media, we developed a new medium (SHI medium) by combining the ingredients of several selected media that can support different subpopulations within the original oral microbial community derived from pooled saliva. DGGE and 454 pyrosequencing analysis showed that SHI medium was capable of supporting a more diversified community with a microbial profile closer to that of the original oral microbiota. Furthermore, 454 pyrosequencing revealed that SHI medium supported the growth of many oral species that have not before been cultured. Crystal violet assay and the confocal laser scanning microscope analysis indicated that, compared with other media, SHI medium is able to support a more complex saliva-derived biofilm with higher biomass yield and more diverse species. This DGGE-guided method could also be used to develop novel media for other complex microbial communities.
© 2010 John Wiley & Sons A/S.
Figures
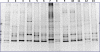



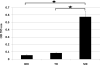
 indicates significant difference (P< 0.05) between the two values.
indicates significant difference (P< 0.05) between the two values.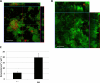
References
-
- Barlaan EA, Sugimori M, Furukawa S, Takeuchi K. Profiling and monitoring of microbial populations by denaturing high-performance liquid chromatography. J Microbiol Meth. 2005;61:399–412. - PubMed
-
- Cook GS, Costerton JW, Lamont RJ. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J Periodontal Res. 1998;33:323–327. - PubMed
-
- Dahlen G. Role of suspected periodontopathogens in microbiological monitoring of periodontitis. Adv Dent Res. 1993;7:163–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

