Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules
- PMID: 20884282
- PMCID: PMC3111150
- DOI: 10.1016/j.mib.2010.08.012
Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules
Abstract
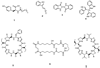
Bacteria grown in pure culture have been the starting point for the discovery of many of the antibacterials now in use. Metagenomics, which utilizes culture-independent methods to access the collective genomes of natural bacterial populations, provides a means of exploring the antimicrobials produced by the large collections of bacteria that are known to be present in the environment but remain recalcitrant to culturing. Both novel small molecule antibiotics and new antibacterially active proteins have been identified using metagenomic approaches. The recent application of metagenomics to the discovery of bioactive small molecules, small molecule biosynthetic gene clusters and antibacterially active enzymes is discussed here.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures


References
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. - PubMed
-
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. - PubMed
-
- Torsvik V, Daae FL, Sandaa RA, Ovreas L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol. 1998;64:53–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

