Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics
- PMID: 20884963
- PMCID: PMC2947301
- DOI: 10.2353/ajpath.2010.100104
Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics
Abstract
Alterations of the p53 pathway are among the most frequent aberrations observed in human cancers. We have performed an exhaustive analysis of TP53, p14, p15, and p16 status in a large series of 143 soft tissue sarcomas, rare tumors accounting for around 1% of all adult cancers, with complex genetics. For this purpose, we performed genomic studies, combining sequencing, copy number assessment, and expression analyses. TP53 mutations and deletions are more frequent in leiomyosarcomas than in undifferentiated pleomorphic sarcomas. Moreover, 50% of leiomyosarcomas present TP53 biallelic inactivation, whereas most undifferentiated pleomorphic sarcomas retain one wild-type TP53 allele (87.2%). The spectrum of mutations between these two groups of sarcomas is different, particularly with a higher rate of complex mutations in undifferentiated pleomorphic sarcomas. Most tumors without TP53 alteration exhibit a deletion of p14 and/or lack of mRNA expression, suggesting that p14 loss could be an alternative genotype for direct TP53 inactivation. Nevertheless, the fact that even in tumors altered for TP53, we could not detect p14 protein suggests that other p14 functions, independent of p53, could be implicated in sarcoma oncogenesis. In addition, both p15 and p16 are frequently codeleted or transcriptionally co-inhibited with p14, essentially in tumors with two wild-type TP53 alleles. Conversely, in TP53-altered tumors, p15 and p16 are well expressed, a feature not incompatible with an oncogenic process.
Figures


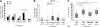


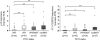

References
-
- Chibon F, Mariani O, Derré J, Malinge S, Coindre JM, Guillou L, Lagacé R, Aurias A. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet. 2002;139:24–29. - PubMed
-
- Chibon F, Mariani O, Mairal A, Derré J, Coindre JM, Terrier P, Lagacé R, Sastre X, Aurias A. The use of clustering software for the classification of comparative genomic hybridization data. An analysis of 109 malignant fibrous histiocytomas. Cancer Genet Cytogenet. 2003;141:75–78. - PubMed
-
- Coindre JM, Mariani O, Chibon F, Mairal A, De Saint Aubain Somerhausen N, Favre-Guillevin E, Bui NB, Stoeckle E, Hostein I, Aurias A. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol. 2003;16:256–262. - PubMed
-
- Derré J, Lagacé R, Nicolas A, Mairal A, Chibon F, Coindre JM, Terrier P, Sastre X, Aurias A. Leiomyosarcomas and most malignant fibrous histiocytomas share very similar comparative genomic hybridization imbalances: an analysis of a series of 27 leiomyosarcomas. Lab Invest. 2001;81:211–215. - PubMed
-
- Fletcher CDM, Unni KK, Mertens F. Pathology and genetics of tumours of soft tissue and bone. IARC Press; Lyon: 2002. World Health Organisation classification of tumours.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

