Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis
- PMID: 20885380
- PMCID: PMC3104474
- DOI: 10.1038/nprot.2010.131
Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis
Abstract
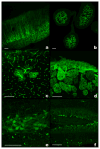
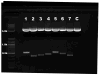
We report here a high-throughput method for the modification of bacterial artificial chromosomes (BACs) that uses a novel two-plasmid approach. In this protocol, a vector modified in our laboratory to hold an R6Kγ origin of replication and a marker recombination cassette is inserted into a BAC in a single recombination step. Temporal control of recombination is achieved through the use of a second plasmid, pSV1.RecA, which possesses a recombinase gene and a temperature-sensitive origin of replication. This highly efficient protocol has allowed us to successfully modify more than 2,000 BACs, from which over 1,000 BAC transgenic mice have been generated. A complete cycle from BAC choice to embryo implantation takes about 5 weeks. Marker genes introduced into the mice include EGFP and EGFP-L10a. All vectors used in this project can be obtained from us by request, and the EGFP reporter mice are available through the Mutant Mouse Regional Resource Center (NINDS/GENSAT collection). CNS anatomical expression maps of the mice are available to the public at http://www.gensat.org/.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Similar articles
-
Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication.Genome Res. 2002 Dec;12(12):1992-8. doi: 10.1101/gr.476202. Genome Res. 2002. PMID: 12466304 Free PMC article.
-
One-Step Bacterial Artificial Chromosome (BAC) Modification: Transfer of the Reporter Vector into BAC/RecA Cells and Selection of Co-Integrates.Cold Spring Harb Protoc. 2020 Jul 1;2020(7):098137. doi: 10.1101/pdb.prot098137. Cold Spring Harb Protoc. 2020. PMID: 32611780
-
One-Step Bacterial Artificial Chromosome (BAC) Modification: Preparation of Plasmids.Cold Spring Harb Protoc. 2020 Jul 1;2020(7):098095. doi: 10.1101/pdb.prot098095. Cold Spring Harb Protoc. 2020. PMID: 32611776
-
Recombineering: a powerful new tool for mouse functional genomics.Nat Rev Genet. 2001 Oct;2(10):769-79. doi: 10.1038/35093556. Nat Rev Genet. 2001. PMID: 11584293 Review.
-
An overview on the generation of BAC transgenic mice for neuroscience research.Curr Protoc Neurosci. 2005 May;Chapter 5:Unit 5.20. doi: 10.1002/0471142301.ns0520s31. Curr Protoc Neurosci. 2005. PMID: 18428622 Review.
Cited by
-
Genomically humanized mice: technologies and promises.Nat Rev Genet. 2011 Dec 16;13(1):14-20. doi: 10.1038/nrg3116. Nat Rev Genet. 2011. PMID: 22179716 Free PMC article. Review.
-
High-resolution characterization of a PACAP-EGFP transgenic mouse model for mapping PACAP-expressing neurons.J Comp Neurol. 2016 Dec 15;524(18):3827-3848. doi: 10.1002/cne.24035. Epub 2016 Jun 3. J Comp Neurol. 2016. PMID: 27197019 Free PMC article.
-
Modified climbing fiber/Purkinje cell synaptic connectivity in the cerebellum of the neonatal phencyclidine model of schizophrenia.Proc Natl Acad Sci U S A. 2022 May 24;119(21):e2122544119. doi: 10.1073/pnas.2122544119. Epub 2022 May 19. Proc Natl Acad Sci U S A. 2022. PMID: 35588456 Free PMC article.
-
Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice.Development. 2015 Nov 15;142(22):3921-32. doi: 10.1242/dev.124271. Epub 2015 Oct 8. Development. 2015. PMID: 26450969 Free PMC article.
-
T cell help shapes B cell tolerance.Sci Immunol. 2024 Feb 16;9(92):eadj7029. doi: 10.1126/sciimmunol.adj7029. Epub 2024 Feb 16. Sci Immunol. 2024. PMID: 38363829 Free PMC article.
References
-
- Monaco AP, Larin Z. YACs, BACs, PACs and MACs: artificial chromosomes as research tools. Trends Biotechnol. 1994;12:280–286. - PubMed
-
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. - PubMed
-
- Ioannou PA, et al. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

