Critical functions of Rpa3/Ssb3 in S-phase DNA damage responses in fission yeast
- PMID: 20885790
- PMCID: PMC2944793
- DOI: 10.1371/journal.pgen.1001138
Critical functions of Rpa3/Ssb3 in S-phase DNA damage responses in fission yeast
Abstract
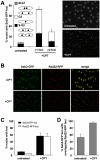
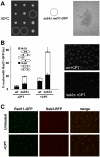
Replication Protein A (RPA) is a heterotrimeric, single-stranded DNA (ssDNA)-binding complex required for DNA replication and repair, homologous recombination, DNA damage checkpoint signaling, and telomere maintenance. Whilst the larger RPA subunits, Rpa1 and Rpa2, have essential interactions with ssDNA, the molecular functions of the smallest subunit Rpa3 are unknown. Here, we investigate the Rpa3 ortholog Ssb3 in Schizosaccharomyces pombe and find that it is dispensable for cell viability, checkpoint signaling, RPA foci formation, and meiosis. However, increased spontaneous Rad11Rpa1 and Rad22Rad52 nuclear foci in ssb3Δ cells indicate genome maintenance defects. Moreover, Ssb3 is required for resistance to genotoxins that disrupt DNA replication. Genetic interaction studies indicate that Ssb3 has a close functional relationship with the Mms1-Mms22 protein complex, which is required for survival after DNA damage in S-phase, and with the mitotic functions of Mus81-Eme1 Holliday junction resolvase that is required for recovery from replication fork collapse. From these studies we propose that Ssb3 plays a critical role in mediating RPA functions that are required for repair or tolerance of DNA lesions in S-phase. Rpa3 orthologs in humans and other species may have a similar function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Mms1-Mms22 complex protects genome integrity in Schizosaccharomyces pombe.DNA Repair (Amst). 2009 Dec 3;8(12):1390-9. doi: 10.1016/j.dnarep.2009.09.008. Epub 2009 Oct 12. DNA Repair (Amst). 2009. PMID: 19819763 Free PMC article.
-
Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe.Genetics. 2007 Sep;177(1):47-61. doi: 10.1534/genetics.107.077255. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660542 Free PMC article.
-
A novel allele of fission yeast rad11 that causes defects in DNA repair and telomere length regulation.Nucleic Acids Res. 2003 Dec 15;31(24):7141-9. doi: 10.1093/nar/gkg917. Nucleic Acids Res. 2003. PMID: 14654689 Free PMC article.
-
The role of MRN in the S-phase DNA damage checkpoint is independent of its Ctp1-dependent roles in double-strand break repair and checkpoint signaling.Mol Biol Cell. 2009 Apr;20(7):2096-107. doi: 10.1091/mbc.e08-09-0986. Epub 2009 Feb 11. Mol Biol Cell. 2009. PMID: 19211838 Free PMC article.
-
S-phase DNA damage checkpoint in budding yeast.Biol Chem. 1998 Aug-Sep;379(8-9):1019-23. doi: 10.1515/bchm.1998.379.8-9.1019. Biol Chem. 1998. PMID: 9792433 Review.
Cited by
-
Comprehensive mutational analysis of the checkpoint signaling function of Rpa1/Ssb1 in fission yeast.PLoS Genet. 2023 May 18;19(5):e1010691. doi: 10.1371/journal.pgen.1010691. eCollection 2023 May. PLoS Genet. 2023. PMID: 37200372 Free PMC article.
-
Fitness profiling links topoisomerase II regulation of centromeric integrity to doxorubicin resistance in fission yeast.Sci Rep. 2015 Feb 11;5:8400. doi: 10.1038/srep08400. Sci Rep. 2015. PMID: 25669599 Free PMC article.
-
Cellular robustness conferred by genetic crosstalk underlies resistance against chemotherapeutic drug doxorubicin in fission yeast.PLoS One. 2013;8(1):e55041. doi: 10.1371/journal.pone.0055041. Epub 2013 Jan 24. PLoS One. 2013. PMID: 23365689 Free PMC article.
-
Unveiling transcriptional mechanisms of B7-H3 in breast cancer stem cells through proteomic approaches.iScience. 2025 Mar 14;28(4):112218. doi: 10.1016/j.isci.2025.112218. eCollection 2025 Apr 18. iScience. 2025. PMID: 40230524 Free PMC article.
-
Tdp1 processes chromate-induced single-strand DNA breaks that collapse replication forks.PLoS Genet. 2018 Aug 27;14(8):e1007595. doi: 10.1371/journal.pgen.1007595. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30148840 Free PMC article.
References
-
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. - PubMed
-
- Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit Rev Biochem Mol Biol. 2009:1–19. - PubMed
-
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. - PubMed
-
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

