CpG-methylation regulates a class of Epstein-Barr virus promoters
- PMID: 20886097
- PMCID: PMC2944802
- DOI: 10.1371/journal.ppat.1001114
CpG-methylation regulates a class of Epstein-Barr virus promoters
Abstract
DNA methylation is the major modification of eukaryotic genomes and plays an essential role in mammalian gene regulation. In general, cytosine-phosphatidyl-guanosine (CpG)-methylated promoters are transcriptionally repressed and nuclear proteins such as MECP2, MBD1, MBD2, and MBD4 bind CpG-methylated DNA and contribute to epigenetic silencing. Methylation of viral DNA also regulates gene expression of Epstein-Barr virus (EBV), which is a model of herpes virus latency. In latently infected human B cells, the viral DNA is CpG-methylated, the majority of viral genes is repressed and virus synthesis is therefore abrogated. EBV's BZLF1 encodes a transcription factor of the AP-1 family (Zta) and is the master gene to overcome viral gene repression. In a genome-wide screen, we now identify and characterize those viral genes, which Zta regulates. Among them are genes essential for EBV's lytic phase, which paradoxically depend on strictly CpG-methylated promoters for their Zta-induced expression. We identified novel DNA recognition motifs, termed meZRE (methyl-Zta-responsive element), which Zta selectively binds in order to 'read' DNA in a methylation- and sequence-dependent manner unlike any other known protein. Zta is a homodimer but its binding characteristics to meZREs suggest a sequential, non-palindromic and bipartite DNA recognition element, which confers superior DNA binding compared to CpG-free ZREs. Our findings indicate that Zta has evolved to transactivate cytosine-methylated, hence repressed, silent promoters as a rule to overcome epigenetic silencing.
Conflict of interest statement
U.R. is one of the founders and shareholders of ChromoTek, which is developing GFP-specific nanobodies for
Figures

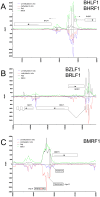
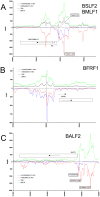
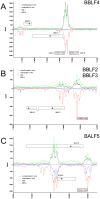

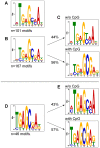


References
-
- Maatouk DM, Kellam LD, Mann MR, Lei H, Li E, et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. - PubMed
-
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. - PubMed
-
- Kieff E, Rickinson AB. Epstein-Barr virus and its replication. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Fields Virology. Philadelphia: Lippincott - Williams & Wilkins; 2007. pp. 2603–2654.
-
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

